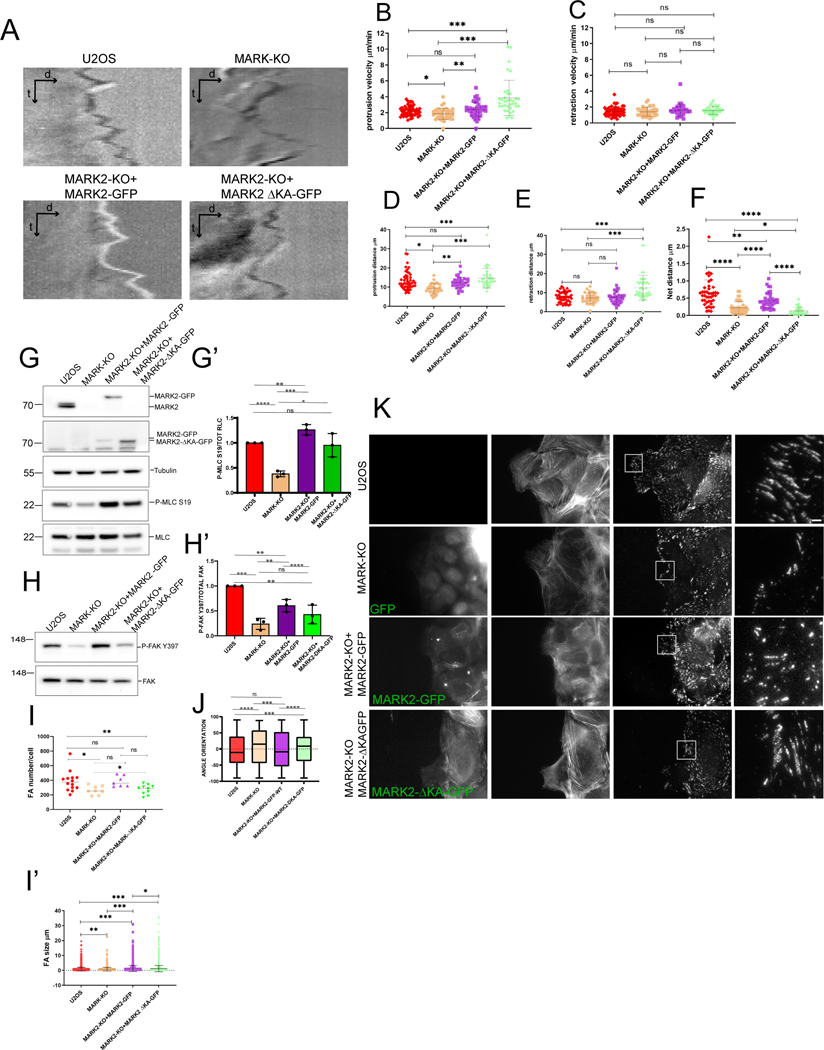
Figure 6: MARK2 association with the plasma membrane promotes FA density and orientation to mediate leading edge advance.
Imaging (A-F, I-K) and western blot (G-H’) of U2OS cells (U2OS), or U2OS cells with MARK2 knocked out by CRSPR-cas9 (MARK2-KO) and MARK3 knocked down by siRNA (MARK-KO) or MARK2-KO cells expressing MARK2-GFP or MARK2-GFP lacking amino acids 1097-1192 (MARK2-ΔKA-GFP). (A) Kymographs generated from time-lapse DIC image series of the leading edges of cells migrating at the edge of a monolayer wound. (B-F) Leading edge dynamics from kymographs. Velocity of protrusion (B) or retraction (C) excursions; distance of protrusion (D) or retraction (E) excursion; (F) Net protrusion distance in a one minute interval. n= 2 kymographs per cell from n= 51 (U2OS), 37 (MARK-KO), 42 (MARK2-KO+MARK2-GFP), 36 (MARK2-KO+MARK2ΔKA-GFP) cells in 2 experiments, bars= means *p<0.01, **p<0.005, ***p<0.0005 ****p<0.0001, Mann-Whitney. (G, H) Western blot of cell lysates probed for MARK2 (G, top), GFP (G, top middle), tubulin (G, middle), myosin regulatory light chain phosphorylated on serine 19 (G, P-MRLC S19, bottom middle) myosin regulatory light chain (G, MRLC, bottom), FAK phosphorylated on tyrosine 397 (H, P-FAK Y397, top) or FAK (H, bottom). (G’, H’) Ratio of P-MRLC S19 to MRLC (G’) or P-FAK Y397 to FAK (H’). n=3 experiments, bars= SEM. (I) Number of FAs per cell (K) for cells migrating at the edge of a monolayer wound. n = 16 (U20S), 9 (MARK-KO), 9 (MARK2-KO+MARK2-GFP) or 10 (MARK2-KO+MARK2ΔKA-GFP) cells in 2 experiments, bars= SD. White box (K) is magnified (ZOOM) at right. Scale bars= 10 μm (left) and 2μm (right). (J) FA orientation relative to the direction of migration in a monolayer wound. n = 5422 (U20S), 2401 (MARK-KO), 3723 (MARK2-KO+MARK2-GFP) or 4041 (MARK2-KO+MARK2ΔKA-GFP) FAs in 2 experiments, box= interquartile range, bar= median, whiskers= upper and lower quartiles. See also Video S7.