FIG 1.
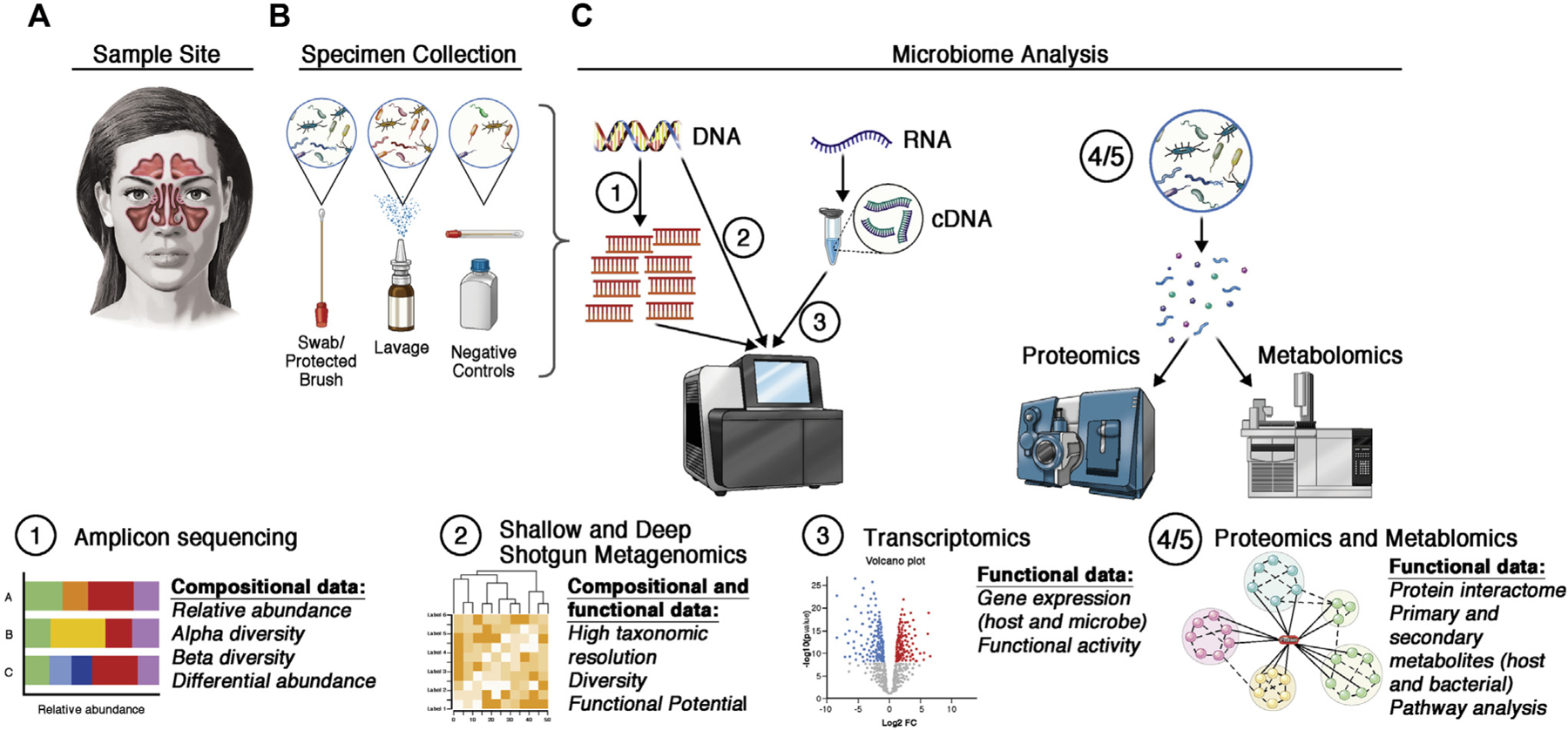
Considerations for analysis of the sinonasal microbiota. A, Investigators must consider the clinical feasibility, benefits, and limitations of the site in the sinonasal cavity. B, Different sampling techniques must also be considered carefully. Negative controls (extraction blanks and blank swabs) can help to identify and reduce contamination. C, Commonly used techniques for microbiome analyses can be used to answer distinct research questions. Amplicon sequencing (eg, 16S rRNA gene sequencing) can yield insights into community composition. Shallow shotgun metagenomics allows for high resolution of the microbial species and strains, whereas deep shotgun metagenomics can yield insights into the putative functions in a community. Gene expression can be analyzed via RNA sequencing (of reverse-transcribed cDNA) to determine which microbial and host genes are expressed in a given condition. Functional analyses can quantify protein production (proteomics) or host- and microbiome-derived metabolites (metabolomics).
