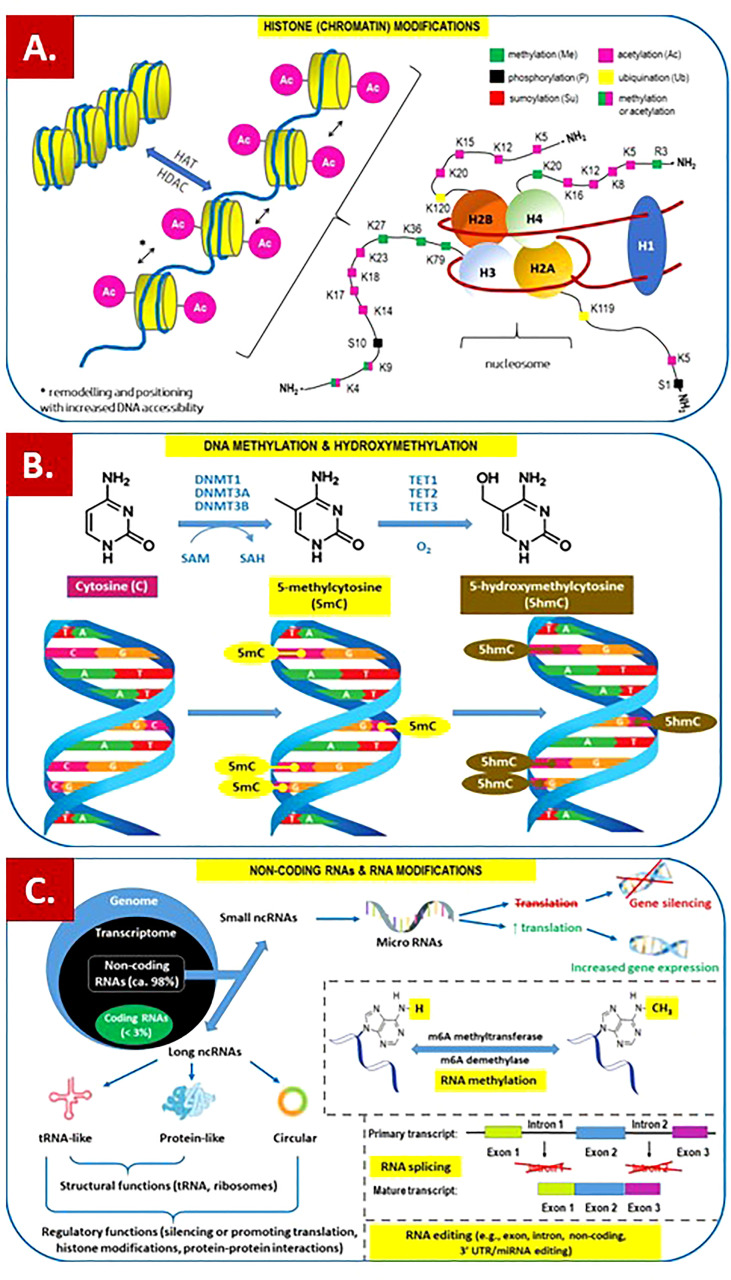
Figure 1.
Overview of the main epigenetic mechanisms that regulate gene expression and may establish potentially heritable changes in gene expression without altering the underlying DNA nucleotide sequence. (A) Histone (chromatin) modifications. On the left. Chromatin remodeling is the dynamic modification of chromatin architecture to allow access of condensed genomic DNA to the regulatory transcription machinery proteins, and thereby control gene expression. For example, histone acetylation by HAT (histone acetyl transferase) increases DNA (chromatin) accessibility because acetylated histones cannot pack as well together as deacetylated histones. HDAC – histone deacetylase; On the right. Each nucleosome consists of two subunits, both made of histones H2A, H2B, H3 and H4, also known as core histones, with the linker histone H1 acting as a stabilizer. Histone post-translational modifications are covalent modifications of histones by phosphorylation on serine or threonine residues, methylation on lysine or arginine, acetylation and deacetylation of lysines, ubiquitylation of lysines and sumoylation of lysines. Histone modifications affect chromosome structure and function, especially during transcription and chromatin remodelling processes. (B) DNA methylation and hydroxymethylation. DNA can be modified at cytosine and adenine residues by the addition of chemical groups. Cytosines can be modified by methylation (5mc) or hydroxymethylation (5hmC), while adenines are modified by methylation. CpG islands (regions of the genome that contain a large number of CpG dunucleotide repeats) are DNA methylations regions in promoters known to regulate gene expression through transcriptional silencing of the corresponding gene. DNA methylation at CpG islands is crucial for gene expression and tissue-specific processes. DNMT – DNA methyltransferase; SAM – S-adenosylmethionine; SAH – S-adenosylhomocysteine; TET – ten-eleven-translocation (methylcytosine dioxygenase). (C) Non-coding RNAs (ncRNAs) and RNA modifications. - ncRNAs play an important role in transcription regulation by epigenetic machinery. Within RNA-induced silencing complexes (RISCs), miRNAs mediate the recognition and binding of RNAs that become targeted for degradation. lncRNAs are associated with other complexes and can activate or repress transcription. - RNA methylation is a post-transcriptional level of regulation. At present, more than 150 kinds of RNA modifications have been identified. They are widely distributed in messenger RNA (mRNA), transfer RNA (tRNA), ribosomal RNA (rRNA), noncoding small RNA (sncRNA) and long-chain non-coding RNA (lncRNA). - Alternative splicing (AS) of pre-mRNAs serves as an additional regulatory process for gene expression after transcription, and it generates distinct mRNA species, and even noncoding RNAs (ncRNAs), from one primary transcript. AS contributes to the diversity of proteins in eukaryotes as cells respond to signals from the environment. AS may lead to generation of ncRNAs, especially long noncoding RNAs (lncRNAs). RNA modifications, such as the RNA N6-methyladenosine (m6A) modification, have been found to regulate AS. - RNA editing is an important mechanism of genetic regulation that amplifies genetic plasticity by allowing the production of alternative protein products from a single gene. RNA editing involves the post-transcriptional insertion and deletion of nucleotides (e.g., uridylate – UMP) within nascent transcripts. RNA editing has been observed in mRNAs, tRNAs, and rRNAs, in mitochondrial and chloroplast encoded RNAs, as well as in nuclear encoded RNAs.