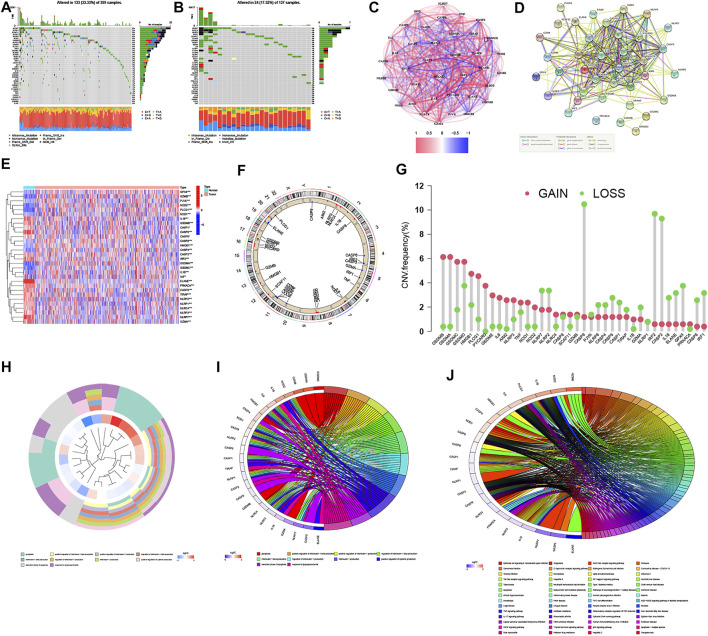
FIGURE 1.
Landscape of the genetic alterations, expressions, and functions of PRGs in CRC. (A,B) Mutation frequencies of 38 PRGs in 399 and 137 patients with COAD and READ based on the TCGA cohort, respectively. (C) Correlation network of PRGs (the depth of the colors reflects the strength of the relevance, red line: positive correlation; blue line: negative correlation). (D) PPI network showing the interactions of PRGs (the minimum required interaction score was 0.4). (E) Heatmap of the PRGs between the normal and the tumor tissues (blue: low expression level; red: high expression level). *p < 0.05; **p < 0.01; and ***p < 0.001. (F,G) Locations of CNV alterations in PRGs on 23 chromosomes and the frequencies of CNV gain, loss, and non-CNV among PRGs, respectively. (H–J) Functional enrichment analysis of the differential expression levels of 30 PRGs ((H): heatmap clustering functional analysis; (I) enriched item in gene ontology analysis; (J) enriched item in Kyoto Encyclopedia of Genes and Genomes analysis).