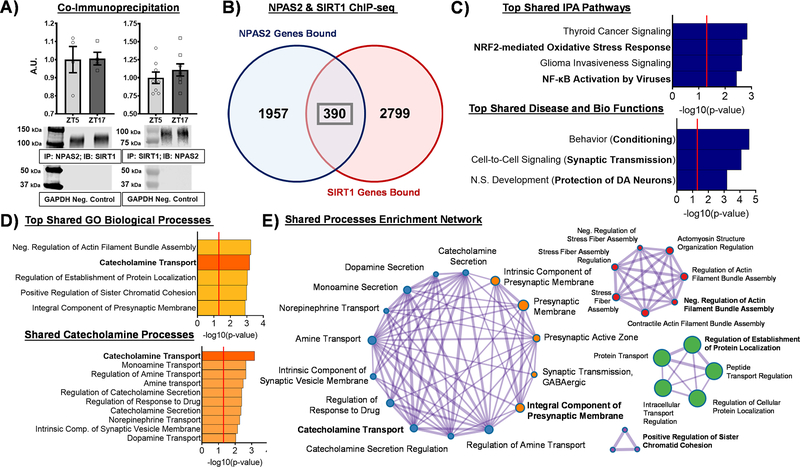
Figure 4. NPAS2 and SIRT1 interact in the NAc and share reward-relevant gene targets.
(A) Co-immunoprecipitation (Co-IP) of NPAS2 from whole NAc tissue and immunoblotting for SIRT1 revealed a band for SIRT1 at ~120kDa across time of day (left), while Co-IP of SIRT1 from whole NAc tissue and immunoblotting for NPAS2 revealed a band for NPAS2 at ~ 92kDa across time of day (right). Data represented as Mean ± SEM (n= 4–8). GAPDH was used as a negative control and was not detected (~37 kDa). (B) Cross analysis of chromatin immunoprecipitation sequencing (ChIP-seq) for NPAS2 and SIRT1 at ZT2, the time shared between the two datasets, revealed 2,347 and 3,189 genes bound (e.g., promoter or gene body) by NPAS2 and SIRT1, respectively, with 390 genes bound in common. (C) The top pathways enriched among these 390 shared genes include metabolic and reward-relevant biological functions. (D) Synaptic transmission, catecholamine transport, and other dopamine-relevant mechanisms are among the top biological processes enriched among NPAS2 and SIRT1’s shared gene targets. (E) Among the enriched shared biological processes, the catecholamine nodes and synaptic transmission nodes show a high degree of interconnectivity. Red line in bar graphs indicates significance threshold of p<0.05 or -log10(p-value) > 1.3.