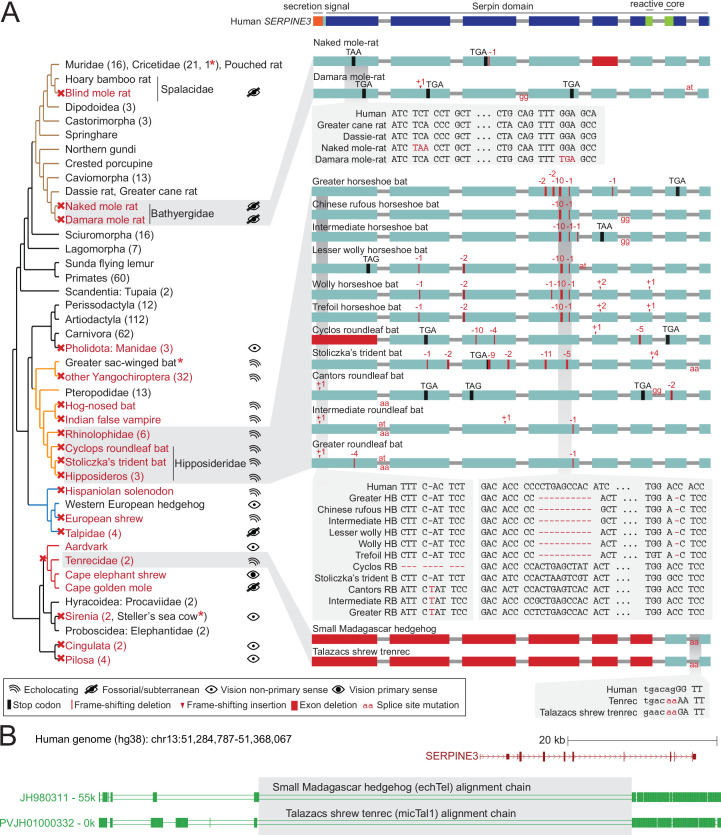
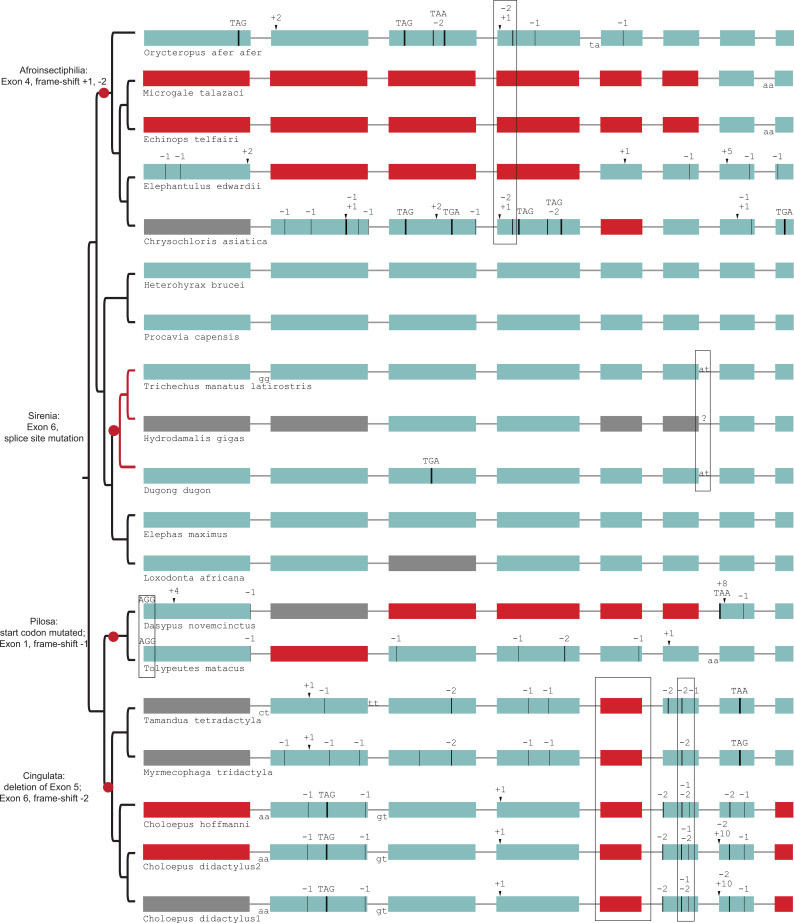
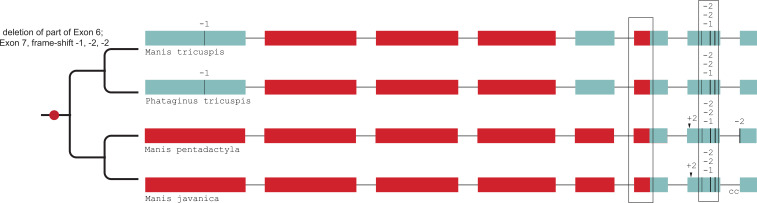
Figure 2. SERPINE3 gene loss pattern across 430 mammalian species.
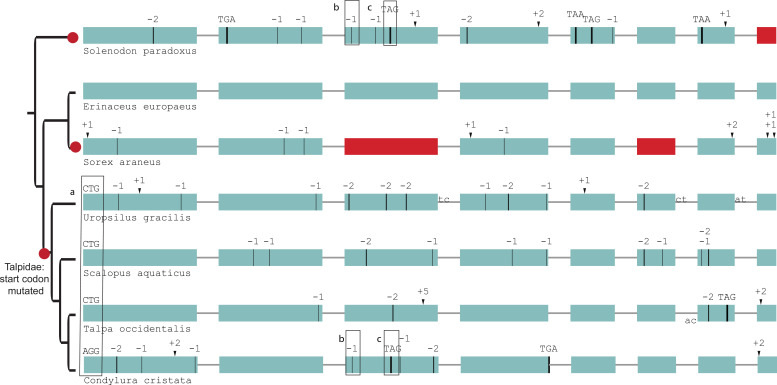
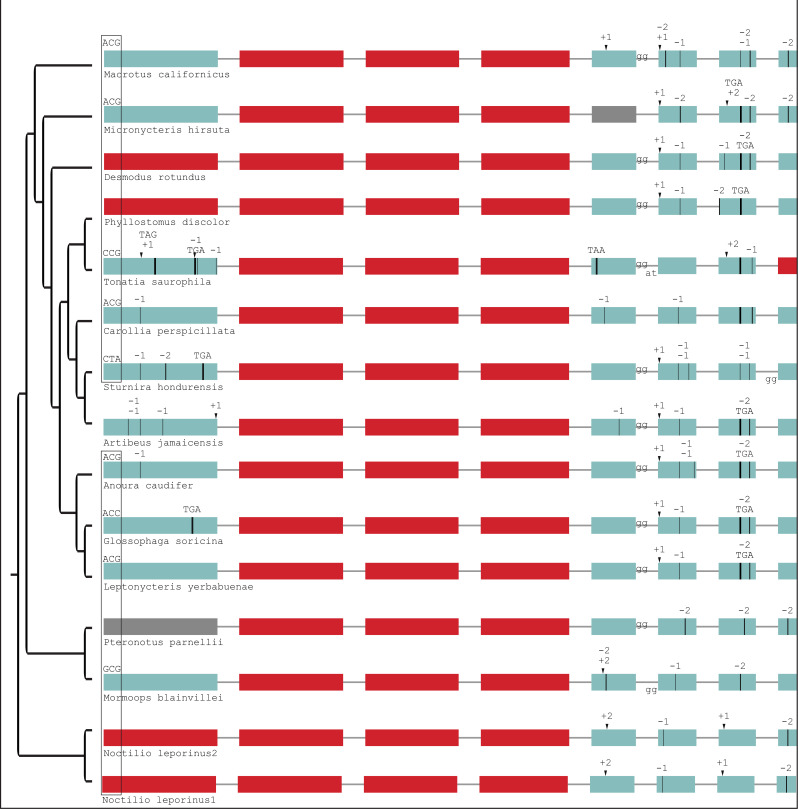
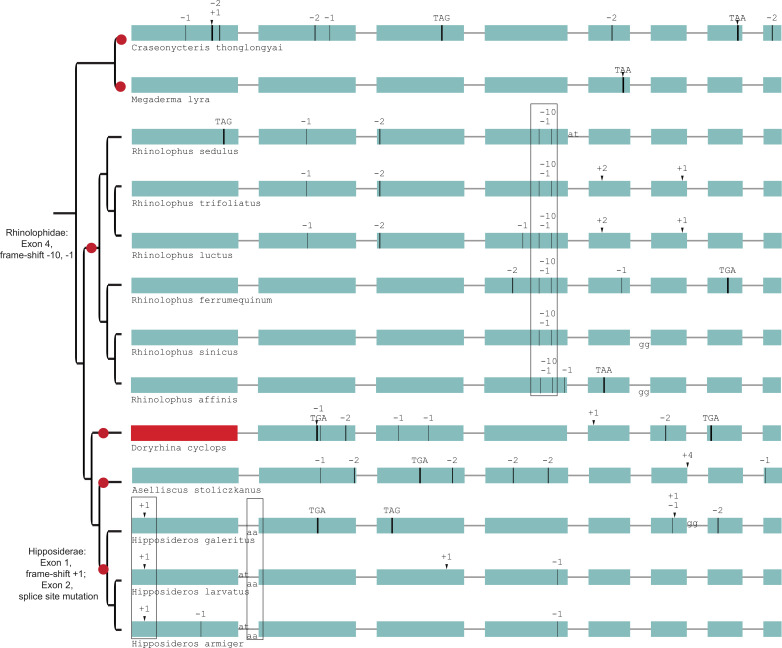
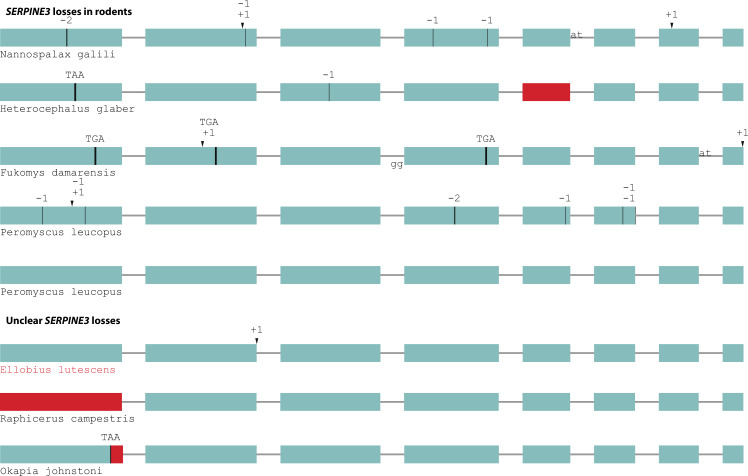
(A) Left: Phylogeny of mammalian species investigated for the loss of SERPINE3 with mapped gene loss events indicated as red crosses. Branches of major clades are colored (Rodentia – brown, Chiroptera – orange, Eulipotyphla – blue, Afroinsectiphilia - red). The number of species investigated per clade is specified in parenthesis. For all loss lineages (red font), visual capability (classified as echolocating, fossorial/subterranean, vision as non-primary and primary sense) is displayed as pictograms at the right. Asterisk marks indicate species, where SERPINE3 evolved under relaxed selection but did not accumulate not more than one inactivating mutation. Right: The Serpin protein domain (Pfam) spans all eight protein-coding exons (boxes) of the intact human SERPINE3 gene (top). The secretion signal and the reactive core region are conserved in species with an intact SERPINE3 (Figure 2—figure supplements 9 and 10). Gene-inactivating mutations are illustrated for three clades, with stop codon mutations shown in black, frame-shifting insertions and deletions shown in red and mutated splice site dinucleotides shown between exons in red. Deleted exons are shown as red boxes. Insets show codon alignments with inactivating mutations in red font. RB - roundleaf bat, HB - horseshoe bat. (B) An ancestral deletion removed large parts of SERPINE3 in the tenrec lineage. UCSC genome browser (Lee et al., 2022) view of the human hg38 assembly showing the SERPINE3 locus and whole genome alignments between human and two tenrec species, visualized as a chain of co-linear local alignments. In these chains, blocks represent aligning sequence and double lines represent sequence between the aligning blocks that do not align between human and tenrec. A large deletion removed the first five protein-coding exons of SERPINE3 in both species. Shared breakpoints (gray box) indicate that the deletion likely represents an ancestral event in Tenrecidae.