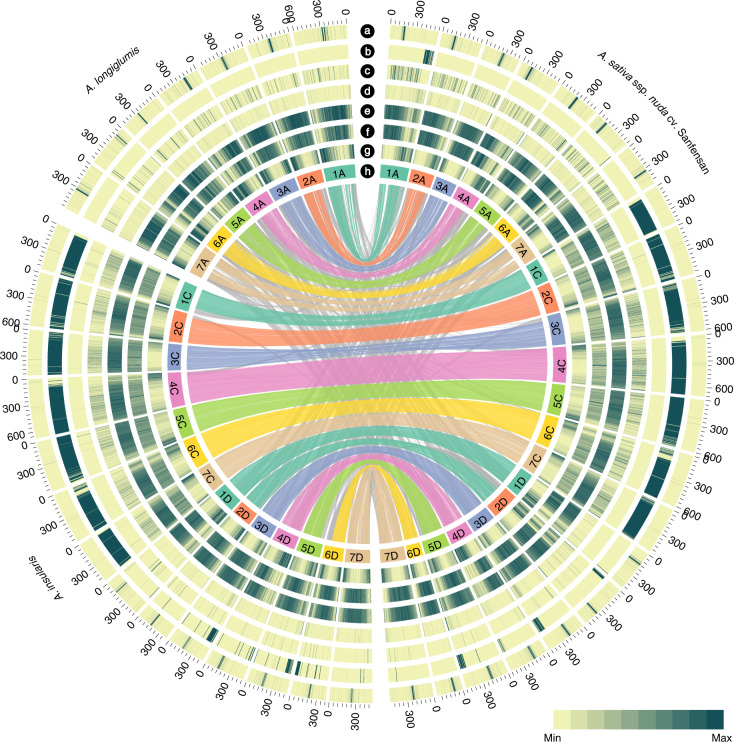
Fig. 1. Circos display of the genomic features of the assembled diploid A. longiglumis, tetraploid A. insularis and hexaploid A. sativa ssp. nuda genomes.
a, The inferred centromere positions of the A and D genome chromosomes. b, The distribution of the C genome-specific repeat Am1 along each chromosome. The Am1-rich regions on chromosomes 1A, 2D, 3D, 4D and 5D of Sanfensan are C genome introgressions. c, The distribution of the A genome-specific repeat As120a along each chromosome. d, k-mer frequencies. e, Tandem repeat (TR) density (MaxPeriod ≤500 bp). f, LTR retrotransposon density. g, Gene density. h, Chromosome names and sizes. The innermost layer shows synteny of hexaploid and its ancestor species, with colored upper-layer links representing syntenic blocks of each hexaploid chromosome and its ancestor chromosome, and the gray low-layer shows chromosome rearrangements during hexaploidization.