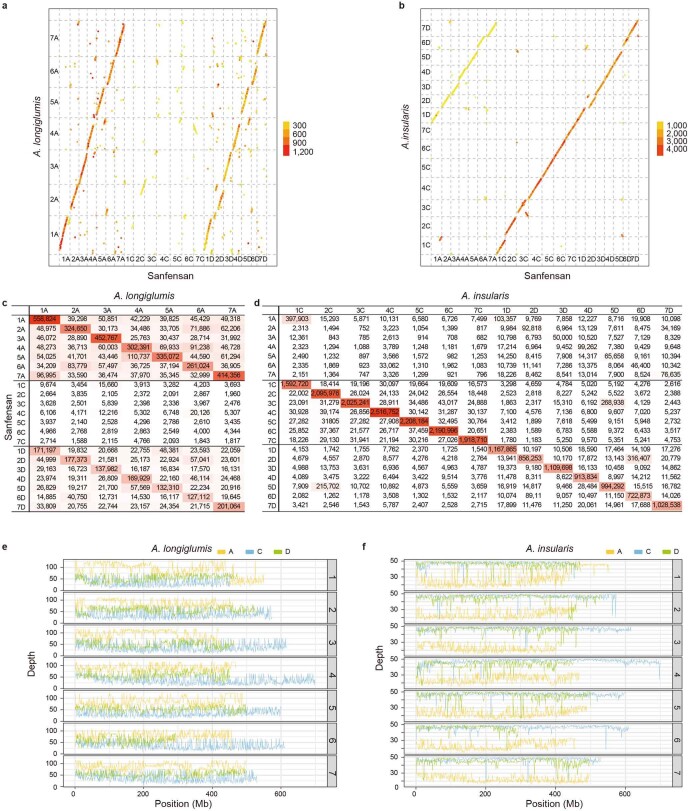
Extended Data Fig. 4. Subgenome assignments in hexaploid oat.
The genome sequences of the diploid A. longiglumis (Al genome) and the tetraploid A. insularis (CD genome) were divided into 100 bp nonoverlapping fragments which were then aligned to the hexaploid ‘Sanfensan’ reference genome. a-b, Dot plots show the distribution of the genomic fragments from A. longiglumis (a) and A. insularis (b) that were uniquely mapped to the ‘Sanfensan’ genome. Each dot represents a syntenic block with at least five syntenic fragments. The distance between each pair of adjacent fragments is <200 kb. c-d, The number of genomic fragments from A. longiglumis (c) and A. insularis (d) that mapped uniquely to the individual ‘Sanfesan’ chromosomes. e-f, Coverage depth obtained along the ‘Sanfensan’ chromosomes after mapping Illumina sequencing reads from A. longiglumis (e) and A. insularis (f) to the ‘Sanfesan’ reference genome.