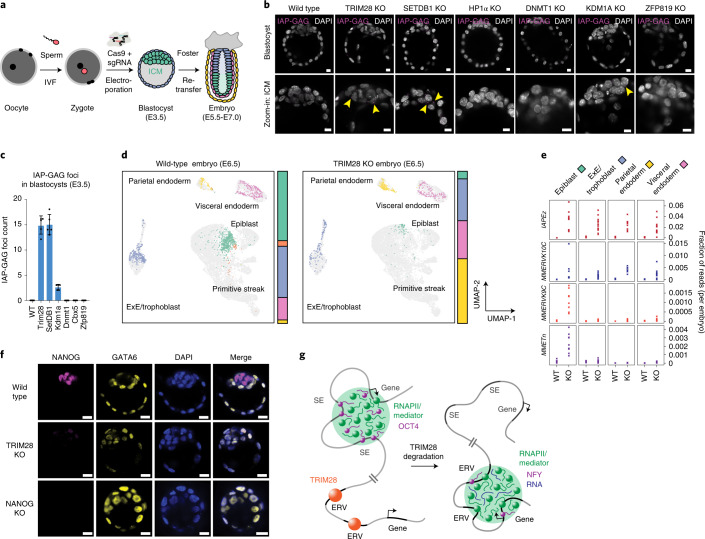
Fig. 5. Early ERV activation correlates with depletion of pluripotent lineages in mouse embryos.
a, Scheme of the zygotic CRISPR–Cas9 perturbation platform. b, IF images of mouse E3.5 blastocysts stained for the GAG protein produced by IAPs. Nuclei are counterstained with DAPI. Note the magenta IAP GAG foci highlighted with yellow arrowheads. Scale bar, 10 µm. c, Quantification of IAP GAG foci in multiple embryos of the indicated genotype across three independent perturbation experiments. Five embryos were picked from the pool of embryos from each genotype for the staining. Each dot represents the GAG foci from an individual embryo. Data are presented as mean values ± s.d. d, Epiblast cells are depleted in TRIM28 KO embryos. Uniform manifold approximation and projection (UMAP) of E6.5 wild-type and E6.5 TRIM28 KO embryos mapped on the combined reference cell state map. The proportions of cells that belong to the individual cell states are indicated as a bar on the right of the UMAP plots. Exe, extraembryonic ectoderm. e, Lineage-specific ERV derepression in TRIM28 KO mouse embryos. The plot shows the fraction of RNA-seq reads that map to the displayed ERV taxa in the indicated cell types in wild-type (WT) and TRIM28 KO embryos in the scRNA-seq data. Each ‘x’ represents a single embryo. f, The inner part of TRIM28 KO blastocysts is populated by GATA6-expressing, NANOG-negative cells. Displayed are representative IF images of NANOG and GATA6 in E3.5 wild-type, TRIM28 KO and NANOG KO blastocysts across two independent perturbation experiments with around 20 embryos per condition. Scale bars, 20 μm. g, Condensate hijacking model. In pluripotent cells, transcriptional condensates associate with SEs bound by pluripotency TFs (for example, OCT4). In the absence of TRIM28, transcriptional condensates are lost from SEs and associate with derepressed ERVs.