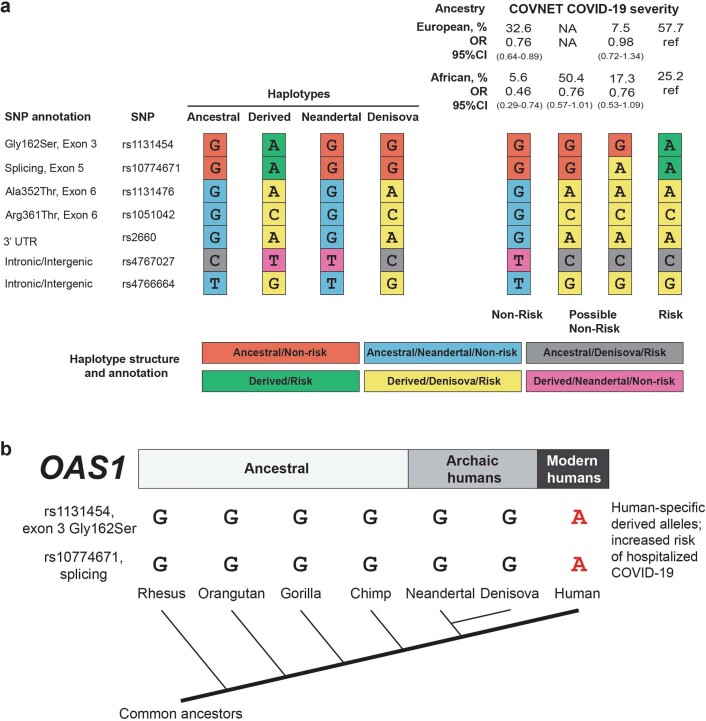
Extended Data Fig. 5. Structure of OAS1 haplotypes in relation to ancestral status and association with COVID-19 severity.
a, Analysis of the OAS1 haplotypes (14.13 kb, hg38:112,911,065-112,925,192) comprised of 7 markers. The color-coding indicates the ancestral status of specific alleles – human (ancestral or derived), archaic humans (Neandertal or Denisova lineages), and COVID-19 severity status (Non-risk/Risk). Haplotype frequencies are shown for hospitalized patients with COVID-19 of European and African ancestry from COVNET. NA, haplotype is not detected. Odds ratios (ORs) and 95% confidence intervals (95%CIs) are for comparison with the common Risk haplotype (also marked as ref); full results can be found in Supplementary Table 4. The Non-risk haplotypes differ from the Risk haplotype by the alleles of rs1131454 and rs10774671. The COVID-19 risk is associated with human-specific derived alleles rs1131454-A and rs10774671-A. Additionally, the Risk haplotype includes a Denisova-type fragment of derived alleles spanning rs1131476 to rs4766664. Human polymorphisms rs1131454, rs1131476, rs1051042, and rs2660 were also explored and found monomorphic in genomic sequences of 29 chimpanzees: Pan troglodytes verus (n = 24) and Pan troglodytes troglodytes (n = 5). Source8: European Nucleotide Archive https://www.ebi.ac.uk, accession numbers FM163403.1–FM163432.1. b, OAS1 haplotypes and phylogenetic tree. COVID-19 risk alleles rs1131454-A and rs10774671-A are human-specific and derived.