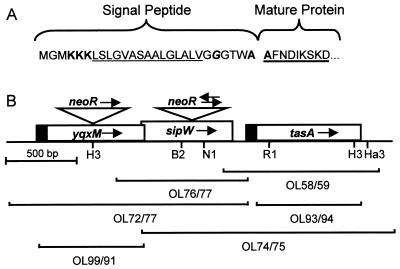
FIG. 2.
(A) N terminus of TasA. The putative signal peptide and the N terminus of the mature protein are indicated. Sequence features consistent with a signal peptide are indicated: three positively charged lysines (bold) close to the N terminus, followed by a stretch of hydrophobic amino acids (underlined) and a glycine residue (italic and bold) 5 amino acids before the possible signal peptidase cleavage site. The two alanines (one at the C terminus of the putative signal peptide, and the other at the N terminus of the mature protein), consistent with a signal peptidase cleavage site, are bold. The sequenced region of the mature protein is heavily underlined. (B) tasA locus. yqxM, sipW, and tasA are indicated by boxes. The solid portions of the boxes encode putative signal peptides. The arrows in the boxes indicate the presumed direction of transcription. The brackets represent the PCR products used in constructing mutations in the locus and for overexpression. The oligonucleotides (Table 2) used to generate each PCR product are indicated. The triangles indicate the disruptions of yqxM and sipW by insertion of the neomycin resistance gene cassette. The arrows above the triangles indicate the directions of transcription of the neomycin resistance genes. Restriction enzyme recognition sites are indicated: B2, BstEII; RI, EcoRI; Ha3, HaeIII; H3, HindIII; NI, NdeI.