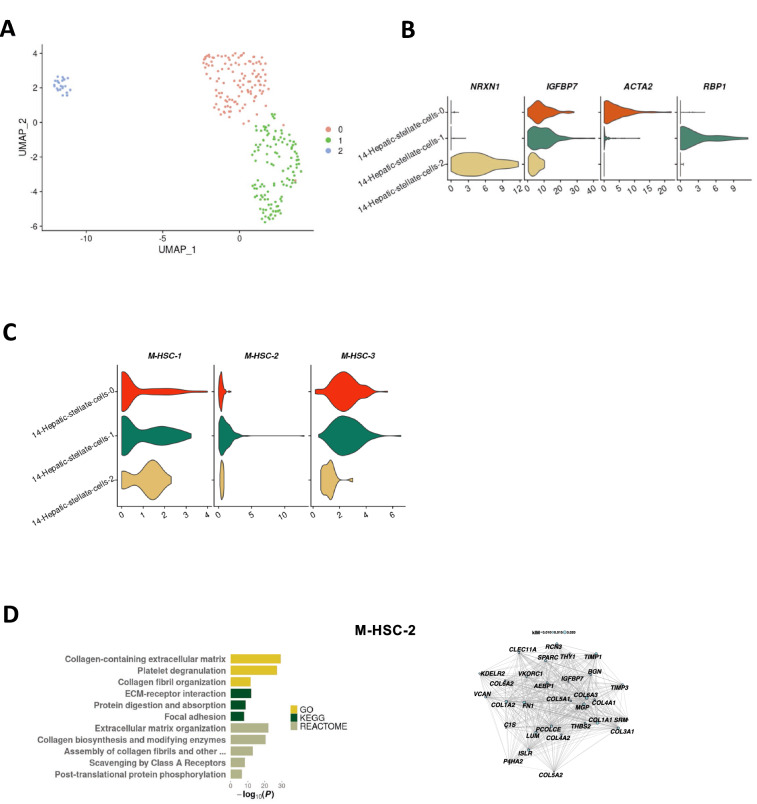
Figure 5.
Activated human hepatic stellate cells are divided in two functionally specific subpopulations. (A) UMAP projection of the three hepatic stellate cell clusters. (B) Violin plots of normalised and batch-corrected expression of hepatic stellate cell marker genes. (C) Violin plots of the activity of hepatic stellate cell modules. (D) M-HSC-2, of which the 30 most central genes are seen in the network diagram. In the network diagram, node sizes represent gene centrality in the module (‘kIM’), and edge widths represent the strength of gene–gene co-expression. For the bar plot, gene sets from the Gene Ontology (GO), Kyoto Encyclopaedia of Genes and Genomes (KEGG) and Reactome databases were queried. Only significant results were plotted (P < 0.05).