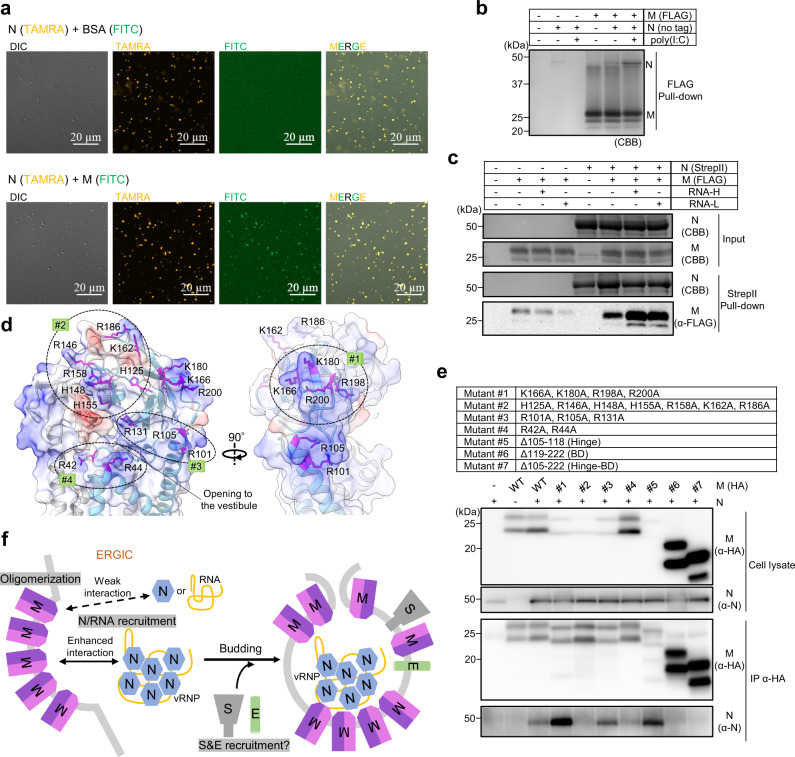
Fig. 4. M protein, N protein, and RNA interactions.
a Differential interference contrast (DIC) and fluorescence images of liquid-liquid phase separation (LLPS) of N protein (TAMRA-labeled) with or without M protein (FITC-labeled). The experiment was repeated three times with similar results. b FLAG tag pull-down assay using recombinant M protein (FLAG-tagged) and N protein (no tag) in the absence or presence of poly(I:C). The experiment was repeated twice with similar results. Source data are provided as a Source Data file. c StrepII tag pull-down assay using recombinant N protein (StrepII-tagged) and M protein (FLAG-tagged) in the absence or presence of RNAs of different sizes. RNA-H and RNA-L indicate yeast RNA with molecular weights >30 kDa and 3–30 kDa, respectively. The experiment was repeated twice with similar results. d Electrostatic surface potentials of the intravirion side of the M protein dimer (long form). Positively charged residues to which the mutations were introduced in e are shown using stick representations and labeled. e Co-immunoprecipitation assay of wild-type (WT) or mutant M proteins with N protein in HEK293T cells. The experiment was repeated three times with similar results. Source data are provided as a Source Data file. f Model of M protein-triggered SARS-CoV-2 assembly M protein in the endoplasmic reticulum–Golgi intermediate compartment (ERGIC) forms dimers in two different conformations that assemble into higher-order oligomers to induce membrane curvature. M protein recruits N and genomic RNA in a cooperative manner. S and E proteins are also recruited to the budding site via an unknown mechanism. Source data are provided as a Source Data file.