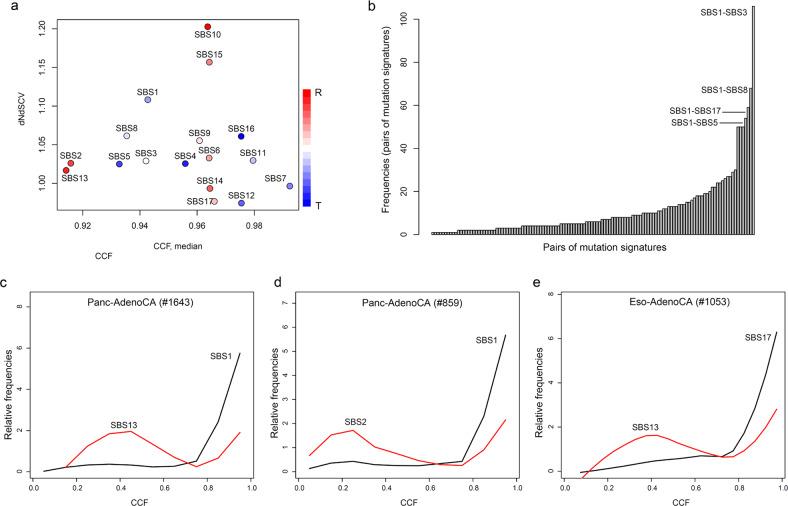
Fig. 4. Evolutionary analysis of mutation signatures.
a Two types of evolutionary measures for mutation signatures are shown in a scatterplot. The cancer cell fraction (CCF, x-axis) and dNdSCV (y-axis) represent levels of mutational clonality and functional selection during cancer evolution, respectively. The dot color indicates the level of replicative and transcriptional strand asymmetries (red and blue, respectively). b For possible pairs of mutation signatures, CCF distributions were compared for concordance using the Kolmogorov–Smirnov test. Significant pairs are shown with their frequencies in a plot highlighting recurrent pairs with SBS1 mutation signatures. The top four most frequent mutation signature pairs are shown. c–e Evolutionary trajectories are shown as relative frequencies of the CCF corresponding to two types of mutation signatures in a single genome. Three example cancer genomes (e.g., two pancreatic adenocarcinomas and one esophageal adenocarcinoma) are shown to illustrate evolutionarily discordant mutation signature pairs, including SBS1-SBS13, SBS1-SBS2, and SBS17-SBS13, which indicate early-late mutations, respectively.