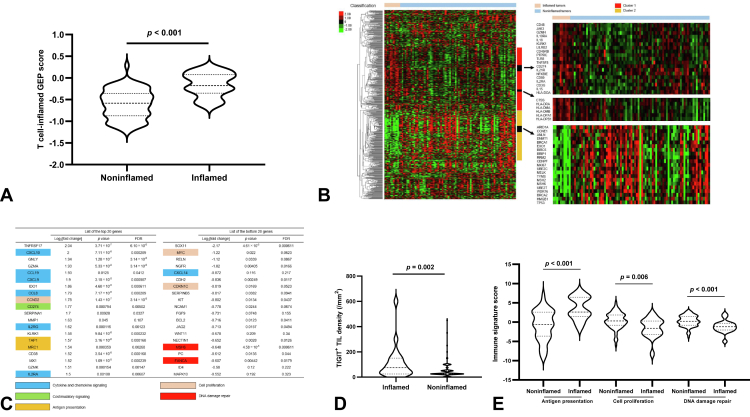
Figure 2.
Transcriptomic features of the tumor immune microenvironment. (A) Violin plots of the T cell–inflamed GEP score for inflamed tumors (n = 17) and noninflamed tumors (n = 72). The p value was determined with the Wilcoxon ranked sum test. (B) Heat map of immune-related gene expression in inflamed tumors (n = 17) compared with noninflamed tumors (n = 72). Each colored square represents the Z score for the expression of one gene, with the highest expression illustrated in red, median in black, and lowest in green. Classification of the tumor immune microenvironment as inflamed or noninflamed is found above the heat map, and expanded views for selected genes of interest in clusters 1 and 2 that were preferentially expressed in inflamed and noninflamed tumors, respectively, are found on the right. (C) List of the top 20 and bottom 20 genes expressed differentially in inflamed tumors relative to noninflamed tumors as determined from volcano plot analysis (Supplementary Fig. 3A). Genes related to antigen presentation, costimulatory signaling, cytokine and chemokine signaling, cell proliferation, or DNA damage repair are shaded as indicated. (D) Violin plots of TIGIT+ TIL density in inflamed (n = 17) and noninflamed (n = 72) tumors. The p value was determined with the Wilcoxon ranked sum test. (E) Violin plots for the expression of gene signatures related to antigen presentation, cell proliferation, or DNA damage repair in inflamed (n = 17) and noninflamed (n = 72) tumors. The p values were determined with the Wilcoxon ranked sum test. FDR, false discovery rate; GEP, gene expression profiling; TIL, tumor-infiltrating lymphocyte.