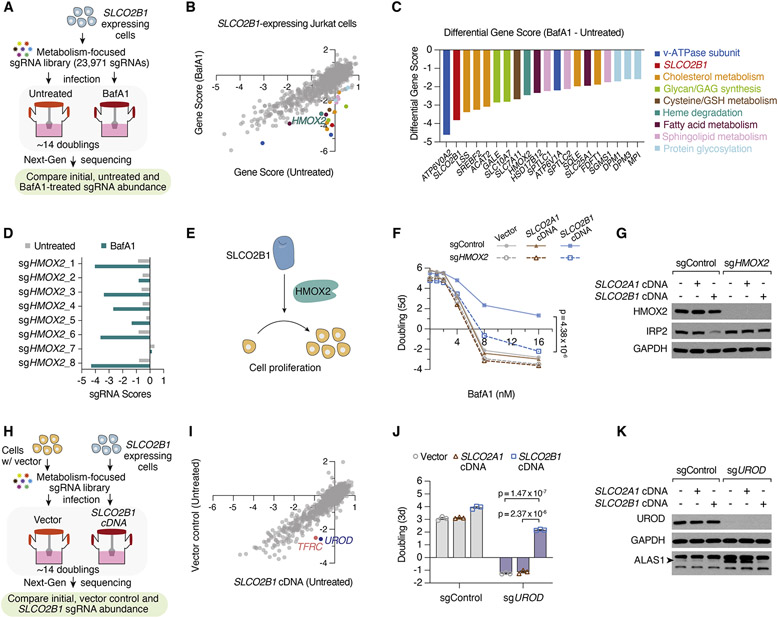
Figure 4. Heme oxygenase is essential for SLCO2B1-mediated resistance to iron restriction.
(A) Schematic describing CRISPR loss of function screens in Jurkat cells expressing SLCO2B1 cDNA in the presence and absence of BafA1.
(B) Gene scores of untreated vs. BafA1-treated (4 nM) Jurkat cells expressing SLCO2B1 cDNA.
(C) Top 20 genes scoring as differentially essential upon BafA1 treatment in SLCO2B1-expressing cells compared to vector controls. Differential gene scores are plotted, and color matched with metabolic processes they are involved in, as listed on right.
(D) sgRNA scores of HMOX2-targeting sgRNAs in untreated and BafA1-treated Jurkat cells expressing SLCO2B1 cDNA.
(E) Schematic illustrating the proposed model for SLCO2B1-mediated rescue of cell proliferation under iron restriction.
(F) Fold change in the number (log2) of control and HMOX2 knockout Jurkat cells expressing a control vector, SLCO2A1 or SLCO2B1 cDNA, during 5-day incubation with or without BafA1 at indicated concentrations. (Mean ± SEM, n= 3, Student’s t-test, 95% confidence interval).
(G) Immunoblot analysis for HMOX2 and IRP2 in control and HMOX2 knockout Jurkat cells expressing vector, SLCO2A1 or SLCO2B1 cDNA. GAPDH was used as loading control.
(H) Schematic depicting CRISPR loss of function screen in Jurkat cells expressing a vector control compared to those expressing SLCO2B1 cDNA.
(I) Gene scores of Jurkat cells expressing a control vector or SLCO2B1 cDNA.
(J) Fold change in the number (log2) of control and UROD knockout Jurkat cells expressing vector, SLCO2A1 or SLCO2B1 cDNA after 3 days. Cells were seeded 7 days after infecting with UROD sgRNA vector. (Mean ± SEM, n= 3, Student’s t-test, 95% confidence interval).
(K) Immunoblot analysis for UROD and ALAS1 in Jurkat cells expressing vector, SLCO2A1 or SLCO2B1 cDNA infected with a control sgRNA or UROD sgRNA. GAPDH was used as loading control. See also Figure S4, Tables S15-S18