Figure 2.
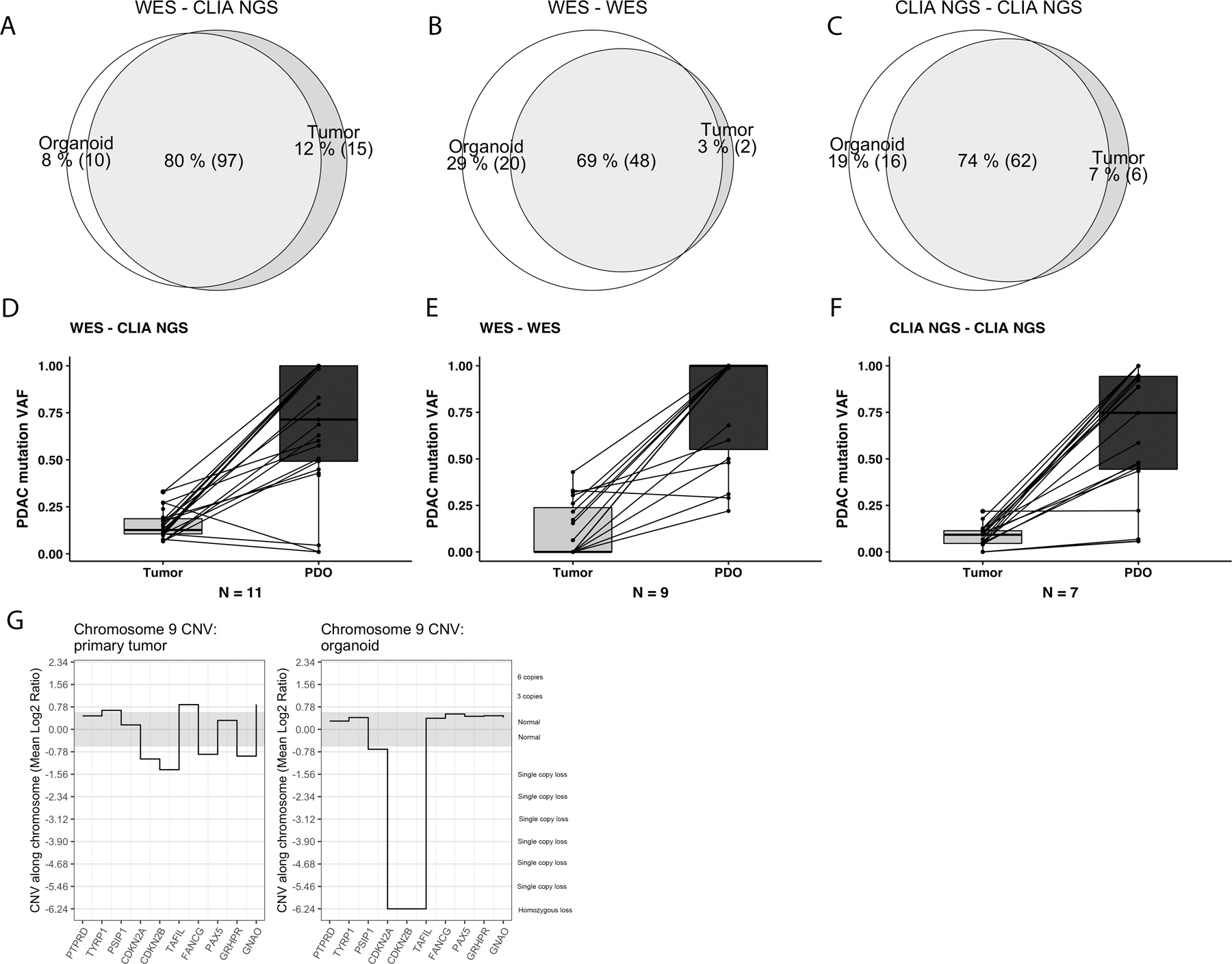
Mutated gene variants detected in three independent datasets by CLIA-approved next generation sequencing of primary tumors and patient-derived organoids (PDO) established from the same primary tumor. In discovery set (A), whole exome sequencing of the PDO was compared to CLIA solid tumor panel (STP) of the primary tumor. In validation set 1 (B), whole-exome sequencing (WES) was performed on both primary tumor and organoid, and in CLIA validation set 2 (C), the CLIA-approved NGS-STP methods were performed for both primary tumor and organoid. The detection of variant allele frequencies (VAF, %) for typical pancreatic cancer driver mutations (KRAS, TP53, SMAD4 and CDKN2A) are substantially higher in PDOs than in primary tumors in discovery dataset (D) and validation datasets 1 (E) and 2 (F). Relevant pancreatic cancer mutations detected by WES of the primary tumor and the corresponding organoids established from the same tumor in the validation set 1 (G). A homozygous copy number loss of the chromosomal area of the CDKN2A gene in the organoid compared to primary tumor. Though demonstrating a slight decrease in CNV, clinically-relevant bioinformatics pipelines would fail to call CDKN2A loss in the primary tumor.
