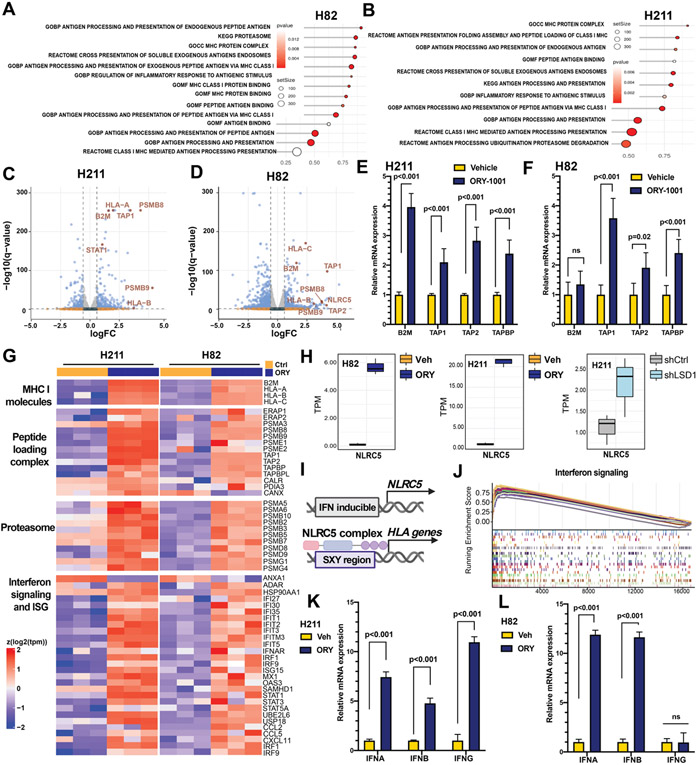
Figure 3: Transcriptomic characterization of SCLC cells after LSD1 inhibition.
Cells treated with 1 μM ORY-1001 or vehicle for 10 days were isolated and subjected to RNA-seq. Pathway enrichment analyses for antigen presentation pathways (APP) are shown as dot plots for (A) H82 and (B) H211. Each dot represents a gene set significantly enriched after LSD1 inhibition. Red indicates significant p values. Dot size reflects number of genes in each gene set. (C-D) Volcano plots showing differential gene expression as beta value (Sleuth-based estimation of log2 fold change) in (C) H211 and (D) H82. APP-related genes are highlighted in red. (E-F) Increased relative mRNA expression of APP-related genes were confirmed by RT-qPCR for (E) H211 and (F) H82. (G) Heatmap reporting mRNA expression of MHC-I molecules, MHC-I peptide complex, proteasomal components and combined interferon signaling and interferon-stimulated genes in H82 and H211 cell lines treated with 1μM ORY ORY-1001 or vehicle (Ctrl). Each column represents an independent replicate. (H) Transcript levels of NLRC5 following LSD1 inactivation in H82 (left; ORY-1001 vs vehicle treatment), H211 (middle; ORY-1001 vs vehicle treatment) and H211 (right; shLSD1 vs shNTC). (I) Schematics showing transactivation of HLA genes by NLRC5 (J) Gene set enrichment analyses for interferon signaling were represented as enrichment scores sorted by rank list. (J-K) Increased relative mRNA expression of interferon genes were confirmed by RT-qPCR for (J) H211 and (K) H82.