Figure 4. Perturbation of rRNA Processing Induces CITIs.
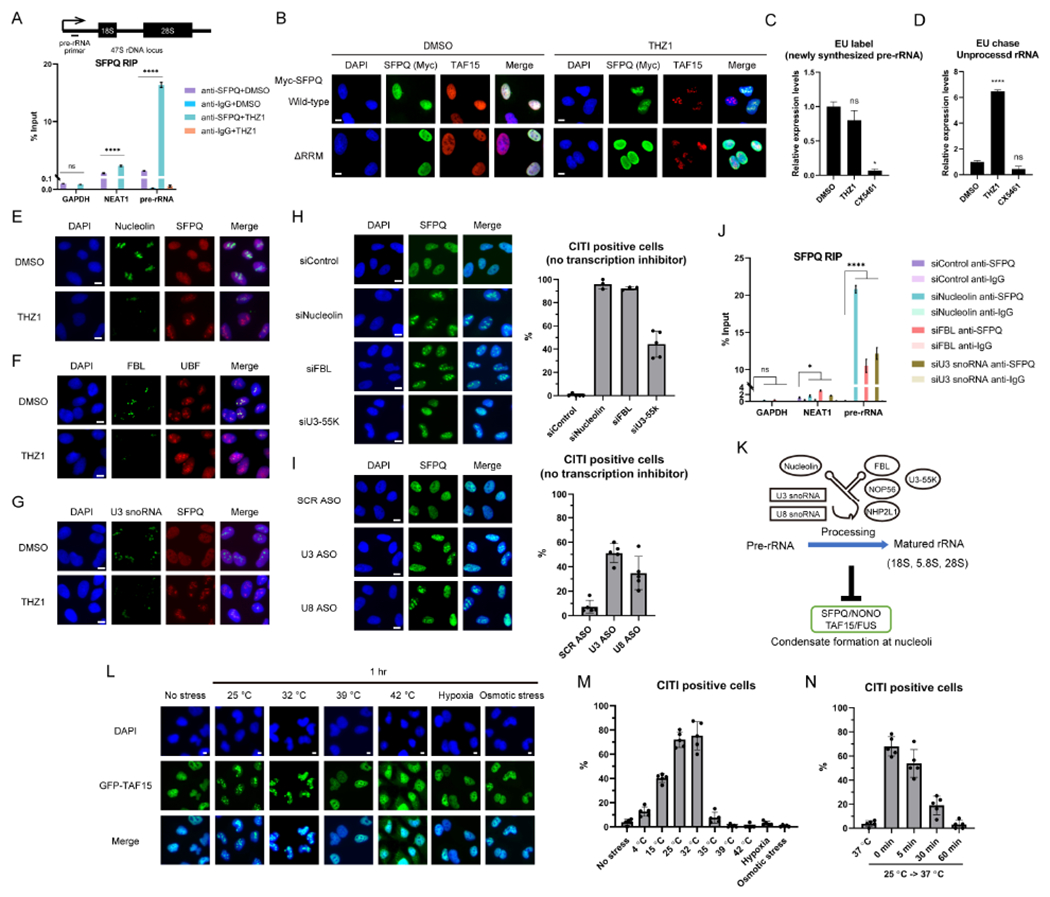
(A) The effect of THZ1 on the rRNA binding of SFPQ was analyzed by RIP (mean with SD, n=3 for measurements from one representative experiment of two biological replicates). The position of the primers for detection of pre-rRNA is shown above.
(B) A SFPQ mutant that lacks RNA-binding domains (ΔRRM) was exogenously expressed and its localization was analyzed by IF. It is of note that the ΔRRM mutant did not localize to CITIs formed by endogenous SFPQ/TAF15. Scale bars, 10 μm.
(C) The newly-synthesized RNA was labeled by EU in the presence of THZ1 or CX5461 for 1 hr. After biotin labeling and pulldown by streptavidin beads, the pre-rRNA levels in EU-labeled RNA were quantified by qPCR (mean with SD, n=3 for measurements from one representative experiment of three biological replicates).
(D) The EU-labeled (1 hr) newly-synthesized RNA was chased for another 1 hr in the presence of THZ1 or CX5461 and analyzed as in (C).
(E-G) The effects of THZ1 treatment on the localization of Nucleolin (E), FBL (F), or U3 snoRNA (G) and CITIs were analyzed by IF. Scale bars, 10 μm.
(H, I) The localization of SFPQ after knockdown (KD) of Nucleolin, FBL, U3-55K (H) or U3, U8 snoRNAs (I) was analyzed by IF. Scale bars, 10 μm. The quantification of CITI positive cells is shown.
(J) The effects of Nucleolin, FBL, or U3 snoRNA KD on the rRNA binding of SFPQ was analyzed as in (A).
(K) A model for the relationship between rRNA processing and condensate formation at nucleoli.
(L, M) The effect of cold/heat shock, hypoxia, or osmotic stress on TAF15 localization was analyzed by IF (L). Scale bars, 10 μm. The quantification of CITI positive cells is shown (M).
(N) The reversibility of CITIs induced by mild cold shock (25 °C) was tested as in Figure 3F.
