Figure 1:
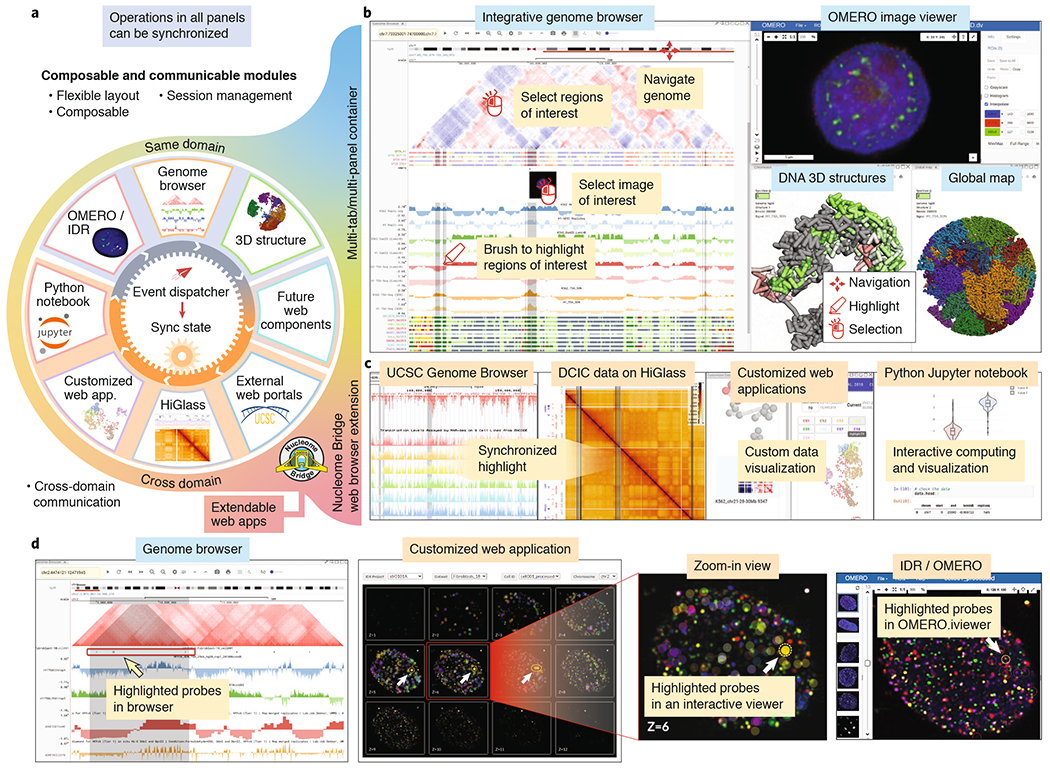
Nucleome Browser facilitates integrative and multimodal data navigation. (a) Overall design of Nucleome Browser. Nucleome Browser consists of a series of web components with synchronized operations, each of which is focused on a specific data type and can be combined in a flexible manner. A unified hierarchical event-dispatch system allows modules (outer circle) from either the same domain or different domains to synchronize their status based on genomic coordinates and highlighted regions. Operations (e.g., navigation to or highlight on the region(s)) initiated in any component will automatically dispatch to all connected web components through the event-dispatch system. Nucleome Bridge, a web browser extension, empowers various external data portals (e.g., the UCSC Genome Browser) to communicate with other modules in the platform. (b) A screenshot of Nucleome Browser illustrates interactive exploration of multimodal 4DN data. Users can use the mouse to navigate to or highlight region-of-interest on 1D and 2D tracks in a genome browser web component. OMERO image viewer shows the details of the image on the selected locus on the image track. 3D genome structure model web component shows a 3D view of chromosome structure with colors indicating the signals of genomic data. (c) The web browser extension named Nucleome Bridge further transmits both genomic coordinates and highlighted regions to other web applications including UCSC Genome Browser, HiGlass viewer, custom data viewers, or Jupyter Notebook to generate plots in real-time. (d) A customized web application can connect Nucleome Browser to the in situ genome sequencing (IGS) data hosted on the IDR platform. Users can navigate the IGS dataset to select an individual cell and this web application will automatically fetch images from IDR and mark the location of probes as circles. As users select regions on the genome browser panel on the left, probes overlapping with the highlighted regions will also be highlighted. Clicking a probe will lead users to the IDR portal and open an OMERO.iviewer for further examination.
