Figure 3. Pfu Cas3 rigidifies the PAM-recognition subunit of Pfu Cascade, enabling DNA target-binding.
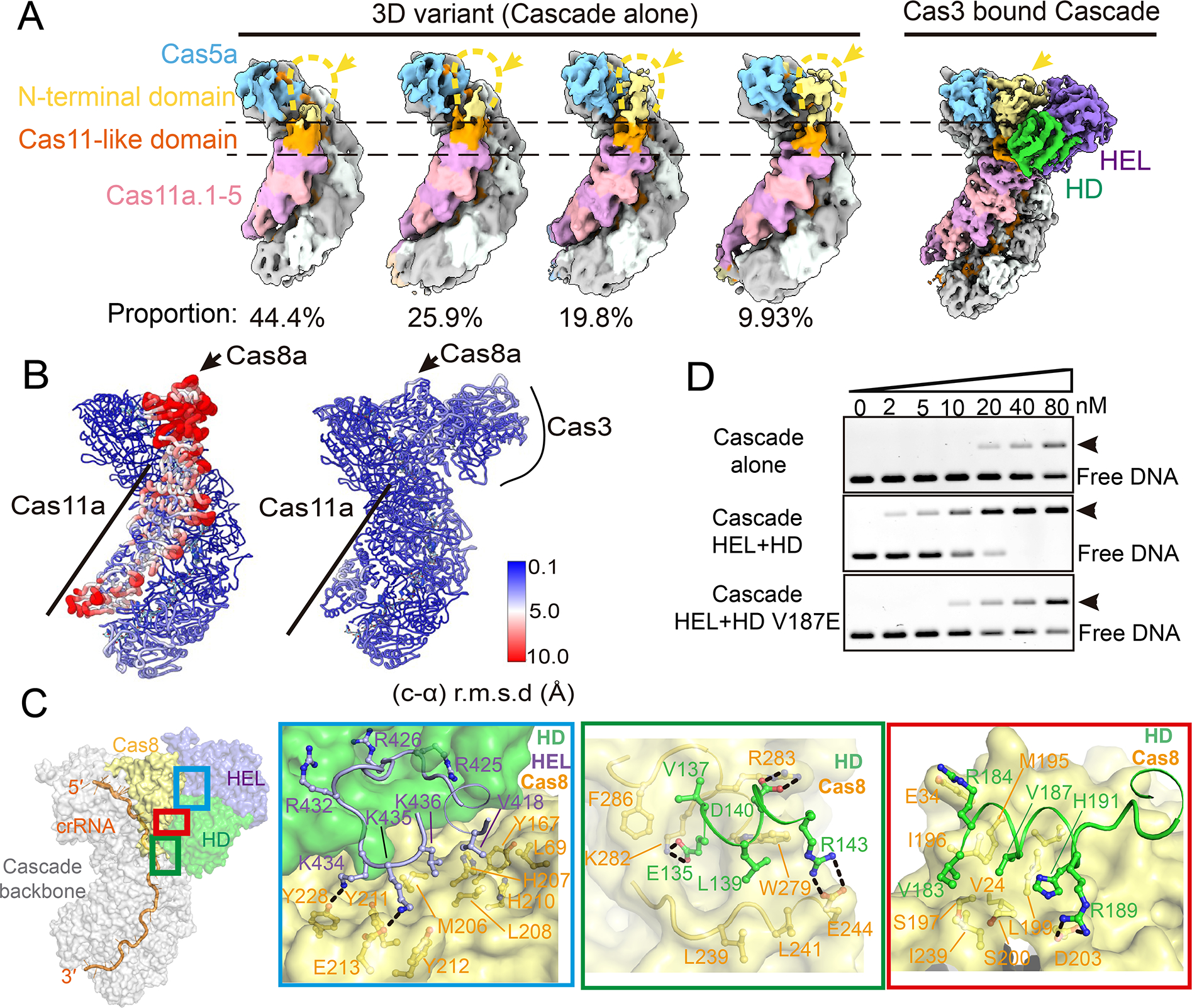
(A) Further classification revealed four 3D variants from the apo Pfu Cascade cryo-EM reconstruction, each represents the specified proportion of the total particles. They vary in the Cas8a NTD density. Only ~10% particles contain choppy densities large enough to cover entire Cas8a. In contrast, Cas8a NTD density is well defined in the Pfu Cascade-Cas3 reconstruction. (B) Local resolution estimate based on the per-residue r.m.s.d. value. Cas8a NTD and the rest of the inner belly subunits in the apo Pfu Cascade have reduced resolution and elevated motion based on this analysis. In contrast, the equivalent subunits in the Pfu Cascade-Cas3 structure are resolved at the same resolution as the rest of the structure. (C) Detailed molecular contacts between Pfu Cas3 and Pfu Cascade. An orientation view is provided to the left. The boxed regions are analyzed in zoom-in panels to the right. (D) Native-agarose EMSA showing that when the molecular contact is disrupted, by the V187E mutation to Cas3 HD, Cas3 can no longer improve the target DNA binding behavior of Cascade as the wild-type Cas3 does.
