Figure 7. I-A PfuCascade-Cas3 causes bi-directional deletion in RNA-guided fashion in human cells.
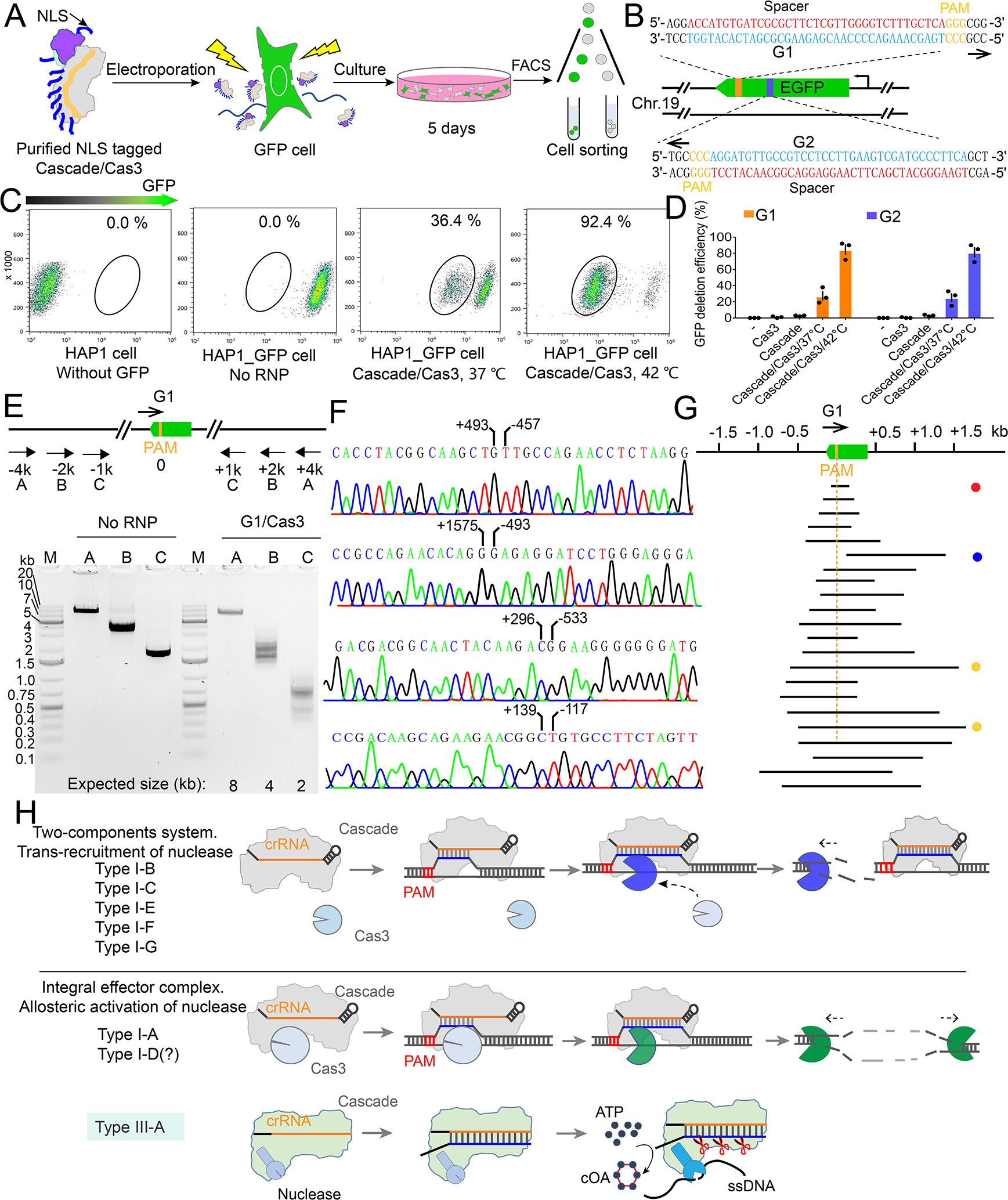
(A) Experimental procedure for Pfu Cascade-Cas3 mediated genome editing in human cells. (B) Design of the crRNA guides targeting the template (G1) and non-template (G2) strands of the GFP ORF. (C) Quantification of the editing efficiency evaluated by FACS, based on the loss of the GFP signal. An overnight incubation at 42°C immediately after RNP delivery increased the editing efficiency from 36.4% to 92.4%. (D) Quantification of G1 and G2 editing efficiency following different experimental procedures. (E) Bracketing PCR based detection and estimation of genome deletion around the targeting site. (F) Representative Sanger sequencing results revealing the deletion boundary, presumably formed by NHEJ-mediated DNA repair. (G) Distribution of the deletion range and size from the Sanger-sequencing results. Note that the vast majority are bi-directional deletions. Target site is eliminated as the result. (H) Mechanism-based classification of Type I CRISPR-Cas systems. I-A and possibly I-D systems use a distinct allosteric activation mechanism to degrade the substrate. These systems may be ancestral to the rest of the Type I systems because they share a stronger mechanistic similarity with Type III systems.
