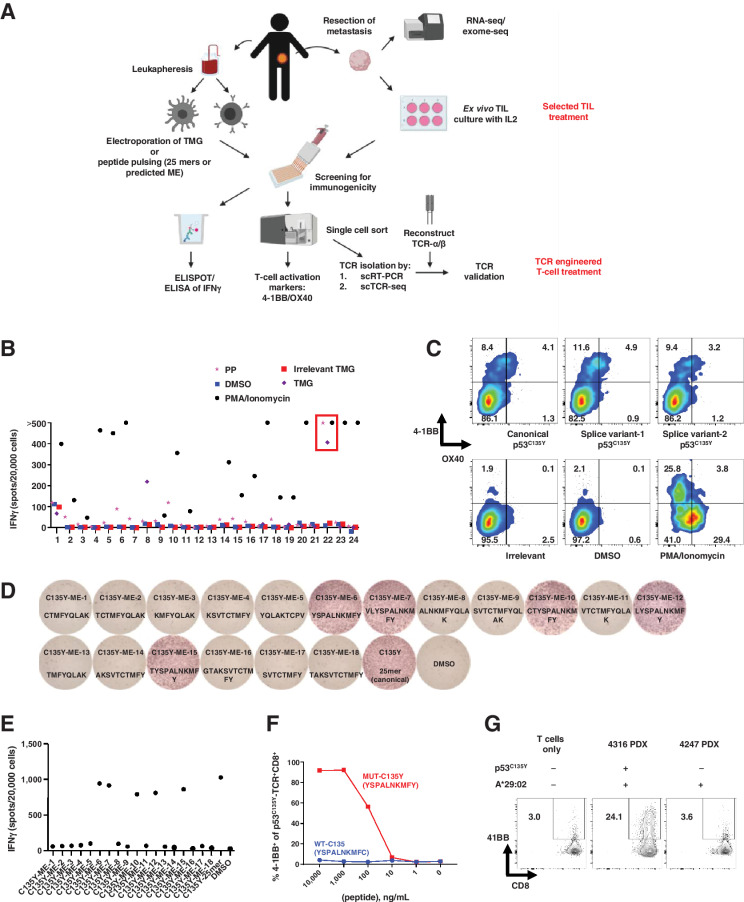
Figure 1.
Unbiased neoantigen screening of TILs from patient 4316 identifies TILs/TCR reactive with mutant p53C135Y. A, Diagram depicting the unbiased neoantigen screening for immunogenicity of shared TP53 mutations. B, ELISpot assay measuring IFNγ secretion in TILs. The 24 TIL fragment subcultures from colorectal cancer patient 4316 were screened against the somatic mutations identified in the patient's tumor, including p53C135Y. TIL fragment 22 showing increased IFNγ secretion against TMGs and PPs that included p53C135Y is highlighted in red (n = 1). C, Flow cytometric analysis of cell surface 4–1BB/OX40 expression upon parsing individual reactivities of TIL fragment 22. An irrelevant peptide (KIAA1328K386R) and DMSO (vehicle) as negative controls and PMA/ionomycin as a positive control are included (n = 1). D, Functional determination of MEs for TCR-B by ELISpot measurement of IFNγ secretion. Eighteen candidate MEs predicted to bind patient's class I HLAs were tested. The amino acid sequences of the tested peptides are listed (n = 1). E, Quantification of IFNγ spots from panel (D). F, Functional testing of avidity of TCR-B. Avidity was determined by coculturing TCR-expressing healthy donor PBLs with autologous imDCs pulsed with serially diluted ME6. 4–1BB expression was measured by flow cytometry (n = 1). G, Autologous tumor cell recognition by 4316 TCR-B. The 4316 autologous PDX was established into a cell line and was cocultured with TCR-B–expressing PBLs. 4–1BB upregulation in p53C135Y-TCR+CD8+ cells is shown. The 4247 PDX line with matching HLA but lacking a p53C135Y mutation was included as a negative control. (n = 1) Experiments in (B) to (G) were independently repeated once. seq, sequencing. PP, peptide pool.