Extended Data Fig. 10. YAP directly induces the transcription of Tead1 and Tead4 in a TET1-dependent manner.
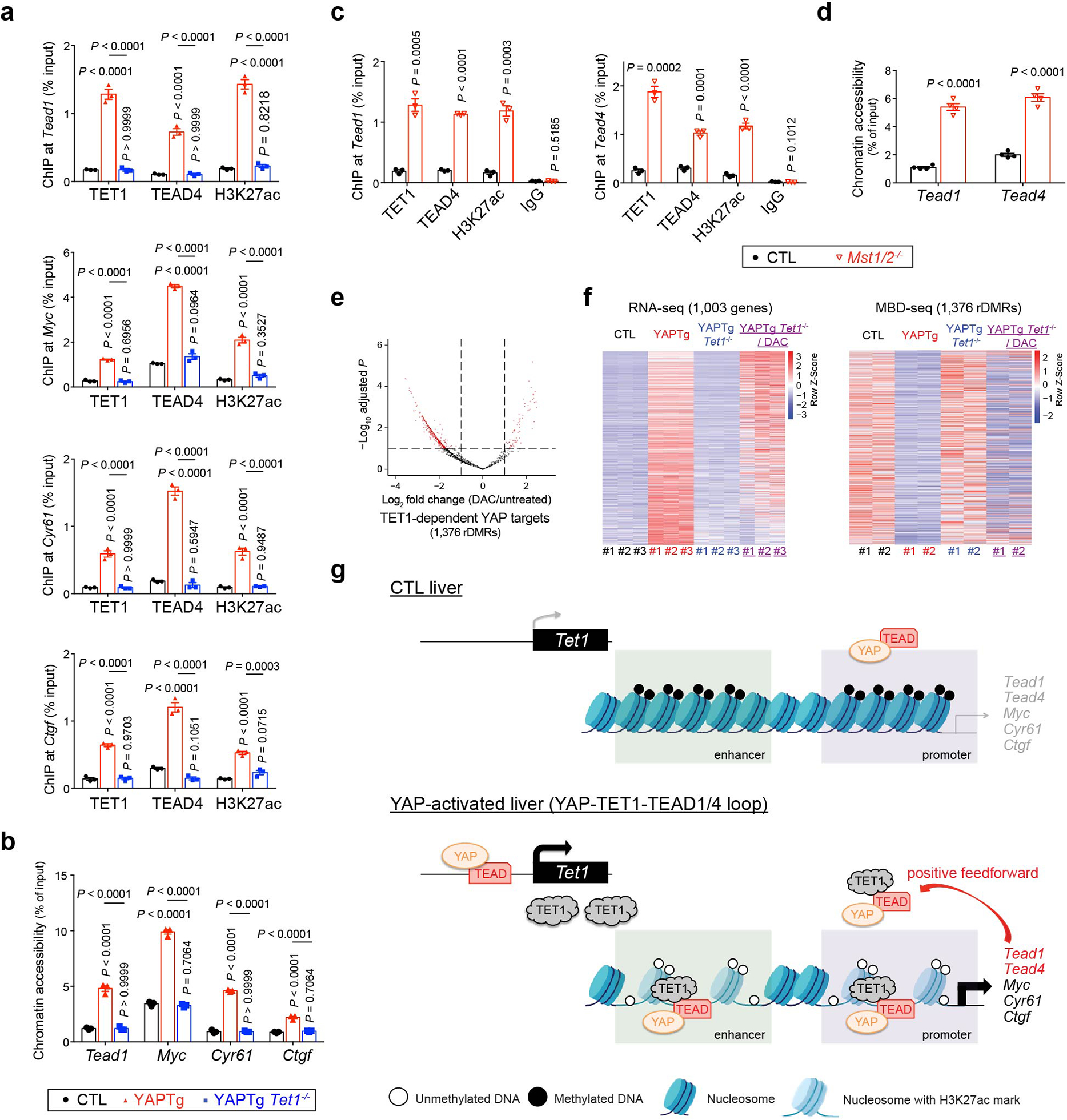
a, b, ChIP-qPCR (n = 3) (a) and FAIRE-qPCR (n = 3) (b) at rDMRs in the indicated mutant livers (also see Fig. 3l and Extended Data Fig. 7e). c, d, ChIP-qPCR (n = 3) (c) and FAIRE-qPCR (n =4) (d) at Tead1 and Tead4 rDMRs in Mst1/2 mutant livers. e, A volcano plot for differential DNA methylation of 1,376 rDMRs sites (associated with 1,003 TET1-dependent YAP target genes) in DAC-treated compared to untreated YAPTg Tet1−/− livers. Red dots represent remarkable (fold change ≥ 2) and significant (P < 0.1) genes. Note that 27.4% of rDMRs (377 out of 1,376) were DNA demethylation by DAC treatment. f, Heatmaps of RNA-seq (left panel) and MBD-seq (right panel) data displaying changes of 1,003 TET1-dependent YAP target genes between the indicated mice, ranked based on P values (between YAPTg Tet1−/− and YAPTg Tet1−/−/DAC) with the most significant on top. g, Schematic model showing a YAP-TET1-TEAD positive feedforward loop sustains YAP signaling. In control livers, the YAP-TEAD complex has limited access to the promoter and enhancer of YAP target genes due to closed chromatin. In YAP-activated livers, YAP-induced expression of TET1 protein causes regional DNA demethylation and local chromatin opening to facilitate YAP-TEAD binding. As a transcriptional coactivator, YAP then recruits histone-modifying enzymes such as NCOA6 and P300, as well as chromatin remodelers such as SWI/SNF complex, to further increase chromatin opening. This further facilitates the association of the YAP-TEAD complex to YAP target genes to activate their transcription. Induction of Tead1 and Tead4 by the YAP-TEAD complex constitutes a feedforward loop to amplify YAP signaling. Values represent mean ± s.e.m. (a, b, c, d). P values are calculated with unpaired two-tailed Student’s t-test (c, d) or one-way ANOVA with Tukey’s test (a, b).
