Fig. 2 |. YAP activation causes regional DNA demethylation in enhancers.
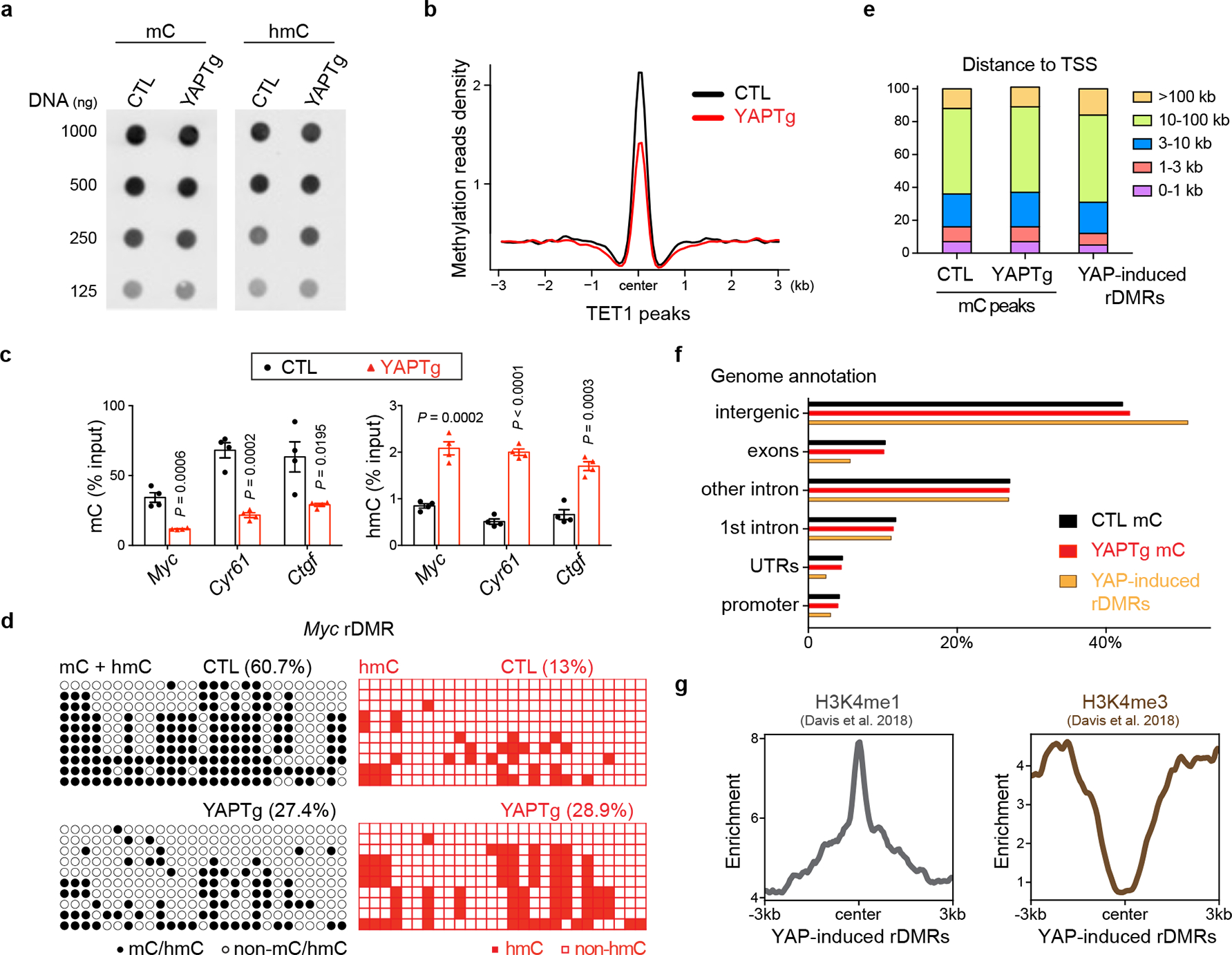
a, DNA dot blot of genomic 5mC and 5hmC levels in YAPTg livers. Images are representative of two independent experiments. b, Methylation profile representing the MBD-seq reads from control and YAPTg livers centered at TET1 peaks. c, 5mC and 5hmC validation at Myc, Cyr61 and Ctgf rDMRs in YAPTg livers. Methylation (left panel) and hydroxymethylation (right panel) levels were determined by MBD-qPCR and hMeDIP-qPCR, respectively (n = 4). Values represent mean ± s.e.m. P values are calculated with unpaired two-tailed Student’s t-test. d, Cloning-based traditional bisulfite sequencing (left panels) and TAB-seq (right panels) of 27 CpG sites at the Myc rDMR locus. All sequencing results include 10 independent clones. e, Absolute distance of methylation peaks and YAP-induced rDMRs relative to nearest TSS. f, Genomic distribution of methylation peaks and YAP-induced rDMRs. YAPTg and control livers showed similar distribution of methylation peaks among intergenic, exon, intron, UTR and promoter at genome-wide level. However, YAP-induced rDMRs were relatively enriched in intergenic (51.0% compared to 42.2%) and depleted in promoter region (3.0% compared to 4.2%). g, Enrichment profiles representing the H3K4me1 ChIP-seq reads (left panel) and the H3K4me3 ChIP-seq reads (right panel) from E15.5 mouse livers centered at YAP-induced rDMRs.
