Fig. 3. Regions of M. capsulatus (Bath) pMMO newly observed in the cryo-EM structures.
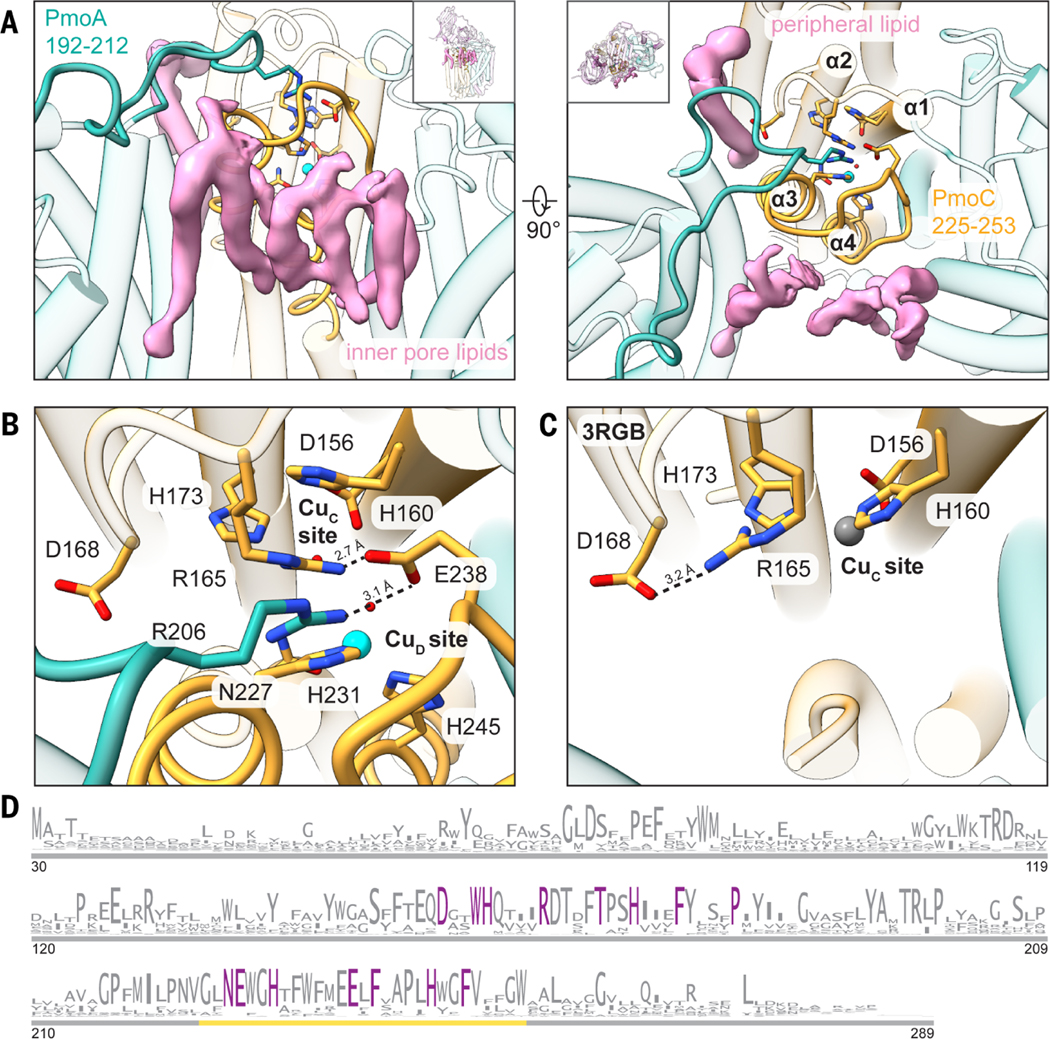
(A) Newly modeled residues in PmoA (192 to 212) (left) and PmoC (225 to 253) (right) are highlighted in teal and gold, respectively. Cryo-EM densities corresponding to lipids that interact with these regions are shown in pink. (B) Magnified view of the completed four-helix bundle from the cryo-EM structure (MC01). Key residues are labeled, and the hydrogen-bonding network involving Arg206 (PmoA), Glu238 (PmoC), and Arg165 (PmoC) is marked with dashed lines. The CuD site copper ion is shown as a cyan sphere. (C) Magnified view of the same region shown in (B) in the crystal structure (Protein Data Bank ID: 3RGB) (9), with the hydrogen bond between Asp168 and Arg165 shown as a dashed line. The zinc ion occupying the CuC site is shown as a gray sphere. (D) Sequence logo for the PmoC subunit (PF04896). Only sequences found in pMMO or ammonia monooxygenase operons are included. Residues conserved at 100% are colored in purple. The gold line demarcates the newly stabilized region in the cryo-EM structures. Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
