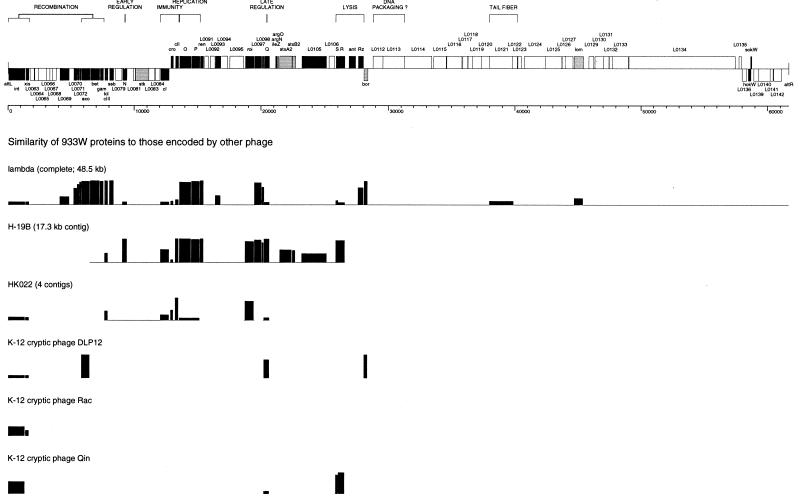
FIG. 2.
Bacteriophage 933W map and comparison to selected other lambdoid phages. (Top) A scale drawing of the molecular map is presented above a base pair scale, arranged in the prophage orientation (attL to attR) as integrated into the E. coli O157:H7 genome. Genes and ORFs with homology to other lambdoid phages are indicated in black. Known or potential virulence factors are indicated in gray (including homologs of the lambda genes lom and bor, members of gene families that have been implicated in virulence in other contexts). All other ORFs, including those with no assigned function, are indicated in white. Gene clusters with related functions, where proposed, are indicated by brackets. (Bottom) Comparison of 933W to other lambdoid phages. Amino acid sequences were aligned with MegAlign (DNASTAR), using the Clustal method with the PAM 250 residue weight table. The percent identity is plotted as a histogram.