Extended Data Fig. 2 |. Single-cell variant calling identifies high-confidence sSNVs.
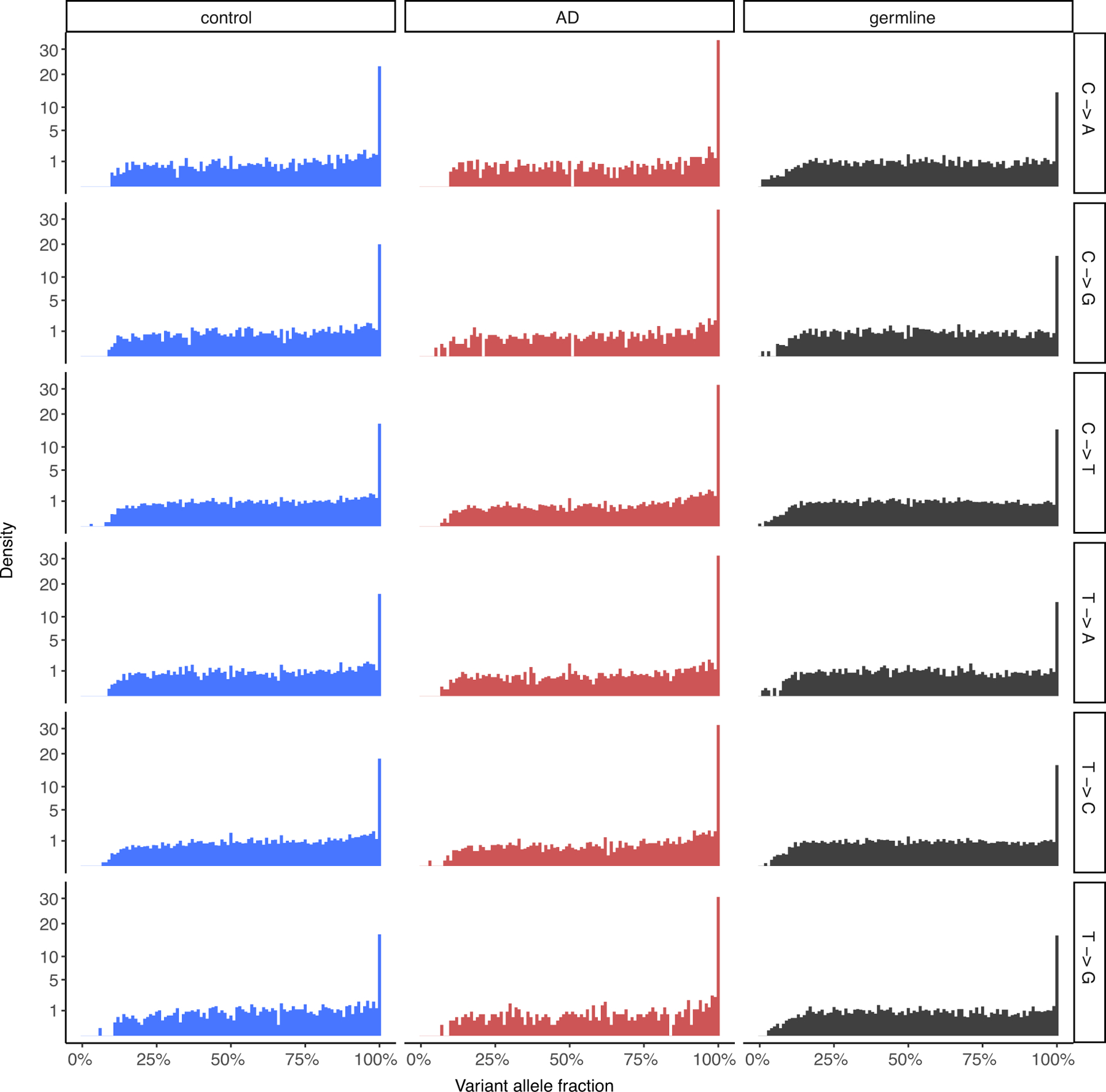
To assess the quality of the sSNVs identified from single-cell MDA-amplified WGS data, we compared their variant allele fractions in control and AD neurons to those of phaseable high-confidence heterozygous germline SNVs from the same neurons, shown for each base change type. The distributions between somatic and germline SNVs are comparable, indicating the validity of the somatic mutation calling method, as has been previously reported for the LiRA calling method5,23.
