Abstract
Liquid biopsy is a novel strategy for tumour diagnosis. The contents of cerebrospinal fluid (CSF) exosomes could reflect glioma status, hence sampling exosomes from CSF is a means of liquid biopsy for glioma. However, few studies have focused on the function of microRNAs in CSF exosomes. In this study, we found that miR‐3184‐3p was enriched in CSF exosomes in glioma patients and was downregulated after tumour resection. We found that miR‐3184 facilitates glioma progression in two ways. On the one hand, miR‐3184 directly promotes proliferation, migration, and invasion while inhibiting apoptosis in glioma. On the other hand, miR‐3184 in glioma‐derived exosomes polarizes macrophages to an M2‐like phenotype, which further aggravates tumour progression. Overall, the current findings uncovered a new mechanism and highlighted the significant role of miR‐3184 in glioma progression. Furthermore, exosomal miR‐3184 could be a considerable factor with potential applications in glioma diagnosis and treatment in the future.
Keywords: exosomes, gliomas, macrophage, microRNAs, tumor immune microenvironment
MiR‐3184‐3p was enriched in cerebrospinal fluid exosomes in glioma patients. MiR‐3184 promotes proliferation, migration, and invasion in glioma. MiR‐3184 in glioma‐derived exosomes polarizes macrophages to an M2‐like phenotype.
Abbreviations
- CSF
contents in cerebrospinal fluid
- IHC
immunohistochemistry
- PDCD4
programmed cell death 4
- RSAD2
radical SAM domain‐containing protein 2
- SAM
S‐adenosylmethionine
- TAMs
tumor‐associated macrophage
1. INTRODUCTION
Glioma is the deadliest malignant brain tumor in adults. 1 Glioblastoma (GBM), which is the most common type of glioma, 2 progresses rapidly, and the median survival for GBM patients is only 14 months. According to the Stupp protocol, tumor resection followed by radiotherapy and temozolomide chemotherapy is the first‐line treatment for GBM patients. 3 However, the effectiveness of this approach is still poor, partially due to a poor understanding of the mechanism of glioma progression.
Tumor‐associated macrophages (TAMs) account for 30% of cells in glioma tissues and display an immunosuppressive effect in glioma. 4 TAMs enhance tumor progression via the secretion of protumor cytokines, exosomes, or anti‐inflammatory factors. 5 , 6 , 7 We previously reported that glioma cells can produce periostin and facilitate the transition of macrophages to the TAM phenotype. 5 This immunosuppressive phenotype resembles that of alternately activated M2 macrophages, therefore we described this process of M2‐like polarization in macrophages. 5 Whether there is another mechanism driving M2‐like polarization in macrophages remains to be studied.
Exosomes are nanoparticles (with diameters between 50 and 150 nm) secreted by a variety of cells, including tumor cells and immune cells. 8 Large quantities of bioactive factors could be transferred cells to cells by exosomes, changing the gene expression in recipient cells. 9 It is believed that exosomes play a key role in mediating tumor progression and the exosomes in plasma and cerebrospinal fluid (CSF) could be used as diagnostic markers for glioma. 10
Aberrant expression of microRNAs (miRNAs) is a hallmark of neoplasms, including glioma. 11 We discovered a series of miRNAs related to glioma proliferation, invasion, mesenchymal transition, and TAM M2‐like polarization. 6 , 7 , 12 , 13 Since exosomal RNA profiles in CSF reflect the disease stages of glioma, 14 we focused on exosomal miRNAs enriched in the CSF of preoperative glioma patients. We proposed that the enriched miRNAs whose expression levels decreased after tumor resection were potential tumor‐related miRNAs. In the current study, we discovered that among these miRNAs, miR‐3184 significantly promoted glioma progression and M2 macrophage polarization, indicating that CSF exosomal miR‐3184 is a potential prognostic factor for glioma.
2. MATERIALS AND METHODS
2.1. Cell lines and cell culture
Human glioma cell lines (LN229 and U251) and THP‐1 cells were obtained from the Culture Collection of the Chinese Academy of Sciences and cultured in Dulbecco's modified Eagle medium (DMEM; Thermo Fisher Scientific) or Roswell Park Memorial Institute 1640 (RPMI 1640; Thermo Fisher Scientific) with 10% fetal bovine serum (FBS; Gibco). The human primary GBM cell line P3 was a kind gift from Dr Rolf Bjerkvig, University of Bergen, and was cultured in neurobasal medium (Gibco) supplemented with GlutaMAX (2 mM), B‐27 supplement (1×), and basic fibroblast growth factor and epidermal growth factor (20 ng/ml). These cell lines were maintained in a humidified chamber containing 5% CO2 at 37 °C.
2.2. Peripheral blood mononuclear cell isolation and induction of macrophages
Peripheral blood mononuclear cells (PBMC) were isolated from the healthy blood of volunteers as previously described. 5 The collected monocytes were cultured in RPMI 1640 complete medium supplemented with 100 ng/ml macrophage colony‐stimulating factor (M‐CSF, Peprotech) for 7 days to induce the M0 macrophage phenotype.
2.3. Patient sample collection
The CSF samples of glioma patients used for exosome miRNA sequencing were isolated from 44 glioma patients who were admitted to Qilu Hospital of Shandong University from November 2017 to October 2019 and the three normal CSF were obtained from shunt procedures of normal pressure hydrocephalus (NPH) patients between February 2018 and December 2018 from the Department of Neurosurgery of the Qilu Hospital of Shandong University. All glioma patients had received surgical resection of gliomas. The study was approved by the Clinical Research Ethics Committee of Qilu Hospital of Shandong University, and written informed consent was obtained from all patients.
2.4. Exosomes labelling
PKH67 (Sigma‐Aldrich) was used to label exosomes as previously described. 15 To eliminate excess dye, PKH67‐labelled exosomes were centrifuged at 100,000 g for 1 h, and the supernatants were discarded. The exosome pellet was diluted in 100 μl of PBS and then used for subsequent uptake experiments.
2.5. Flow cytometry
To detect CD11b+ CD163+ macrophages, anti‐CD163‐PE (BD Biosciences) and anti‐CD11b‐APC (eBioscience) antibodies were used to stain cells according to the manufacturer's instructions. Cells were harvested, washed, and incubated in blocking buffer; isotype controls were run in parallel. Flow cytometry was performed using a BD Accuri C6 flow cytometer (BD Biosciences).
2.6. MiR‐3184‐3p/PDCD4/RSAD2 overexpression and knockdown
Small interfering RNA (siRNA) for PDCD4/RSAD2, PDCD4/RSAD2‐overexpressing vector, and empty vector were purchased from GenePharma. Lentiviruses encoding miR‐3184‐3p mimics, miR‐3184‐3p inhibitor (sh‐miR‐3184‐3p), negative control (NC), and inhibitor negative control (sh‐miR‐NC) were purchased from GeneChem. Transient cell transfection was performed using Lipofectamine 3000 reagent (Thermo Fisher Scientific) according to the manufacturer's protocol. Transfection of the glioma cell lines with lentivirus was performed with a multiplicity of infection (MOI) of 10.
2.7. Exosome isolation and identification
Cerebrospinal fluid from glioma patients in Qilu Hospital of Shandong University was collected and stored in liquid nitrogen. The culture supernatant was collected from glioma cells in DMEM supplemented with 10% exosome‐depleted FBS for 48 h. CSF and glioma cell culture medium were centrifuged at 2000 g for 30 min and 12,000 g for 45 min and filtered through a 0.22‐mm filter to remove cell debris and large vesicles. The collected supernatant was centrifuged at 110,000 g for 70 min. After the supernatant was removed, the pellet was washed with PBS and centrifuged at 110,000 g for 70 min. The exosomes were collected and resuspended in 20–50 ml of PBS for further study.
The exosomes from glioma cells were isolated from conditioned medium from glioma cell lines, and isolation was performed as previously described. 16 The exosomes were stored at −80 °C and verified by electron microscopy and nanoparticle tracking technology (Nanosight).
2.8. Luciferase reporter assay
The reporter genes containing pGL3‐PDCD4/RSAD2 and pGL3‐mut PDCD4/RSAD2 were synthesized by BioAsia. LN229 cells or THP‐1‐derived macrophages were cotransfected with luciferase reporters and miR‐3184‐3p mimics/inhibitors. Forty‐eight hours later, the activity of the reporter protein was measured using a luciferase assay kit (Promega) according to the manufacturer's instructions.
2.9. Western blotting
The harvested cells were lysed using heat denaturation in RIPA cell lysis buffer. The protein lysates were loaded and separated using SDS‐PAGE and then transferred to PVDF membranes. The blots were incubated with primary antibodies targeting the following proteins which can see in Table S1. To visualize the protein bands, enhanced chemiluminescence (ECL; Millipore) was used. The intensity of the protein bands was analysed using ImageJ software and normalized to that of the GAPDH bands.
2.10. Quantitative real‐time PCR
Total RNA was isolated from glioma cells or macrophages using TRIzol reagent (Invitrogen, Life Technologies). Reverse transcription was performed using 2 μg of total RNA and a High‐Capacity cDNA Reverse Transcription Kit (Toyobo, FSQ‐101) according to the manufacturer's protocol. The cDNA was subjected to real‐time PCR using the Mx‐3000P Quantitative PCR System (Stratagene). The primers are listed in Table S2. The mRNA expression was normalized to that of GAPDH to obtain the relative expression level.
2.11. Intracranial mouse model
To establish intracranial gliomas, U251 luciferase cells (1 × 106) transfected with lenti‐miR‐3184‐3p, lenti‐Sh‐miR‐NC, or lenti‐Sh‐miR‐3184‐3p virus were stereotactically implanted into the brains of 4‐week‐old nude mice (SLAC Laboratory Animal Center). For studies testing the interaction between gliomas and macrophages, U251 luciferase cells (1 × 106) mixed with macrophages (2 × 105) transfected with miR‐3184 or miR‐NC (M‐miR‐3184‐3p) were co‐implanted into the brains of nude mice. Bioluminescence imaging was used to detect intracranial tumor growth. Kaplan–Meier survival curves were plotted to determine the survival time. When the mice were moribund or cachectic, the tumor tissues were harvested, fixed in formalin, embedded in paraffin, cut into sections, and stained with hematoxylin and eosin (HE). All procedures that involved mice were approved by the Animal Care and Use Committee of the Qilu Hospital of Shandong University.
2.12. Statistical analysis
Data analysis was performed and data were visualized using GraphPad Prism. Each experiment was carried out at least in triplicate, and all results are presented as the mean ± SD. One‐way ANOVA was used to assess statistical significance. Kaplan–Meier survival curves were also constructed, and log‐rank tests in GraphPad Prism software were used to assess survival. Differences with the following p values were considered significant: P value <0.05, denoted by “*”; P value <0.01, denoted by “**”; P value <0.001, denoted by “***”; and P value <0.0001, denoted by “****.” Difference with P values >0.05 were considered nonsignificant and are denoted by “ns”.
3. RESULTS
3.1. miR‐3184‐3p is enriched in glioma CSF‐derived exosomes and promotes malignancy in glioma cells
First, we examined the exosomal miRNA expression profile in the CSF of glioma patients pre‐ (pre‐CSF) and post‐ (post‐CSF) tumor resection. We found that 123 miRNAs were downregulated after surgery (Figure 1A, pre‐CSF vs. post‐CSF), suggesting that these miRNAs are probably important for glioma maintenance and diagnosis. In addition to fold change, the absolute expression abundance of miRNAs in exosomes should be considered. We therefore ranked the top 20 most highly expressed miRNAs in pre‐CSF exosomes (Figure 1A, exosomal miR) and found that four of these 20 miRNAs showed decreased levels after tumor resection. We further performed miRNA sequencing for glioma tissues and normal brain tissues. The 279 miRNAs upregulated in glioma tissues were selected (Figure 1A, upregulated miR). The intersection of the abovementioned three groups of miRNAs is shown in a Venn diagram, and we finally chose the novel miRNA miR‐3184‐3p for further study (Figure 1A). We further performed nuclear‐cytoplasmic fractionation as well as FISH experiments in four glioma tissues and demonstrated that mir‐3184‐3p is localized in the cytoplasm (Figure S1A,B). We then performed CCK8 and EDU assays and confirmed that proliferation was increased after miR‐3184 overexpression in glioma cells (Figure 1B–D). Overexpression of miR‐3184 in glioma cells also decreased the apoptosis rate (Figure 1E,F). In addition, the migration and invasion abilities of glioma cells increased after miR‐3184 overexpression (Figure 1G‐K). We further silenced miR‐3184 in glioma cells using miR‐3184 inhibitors. We found that miR‐3184 knockdown led to significant decreases in cell viability, invasion, and migration, accompanied by an increase in the cell apoptotic rate (Figure S2A–J). The protumor effect of miR‐3184 in xenografted glioma models in nude mice was examined. The results showed that overexpressing miR‐3184 increased the tumor burden in nude mice, whereas miR‐3184 inhibited tumor growth was silenced (Figure 1L‐M, Figure S2K‐L). Moreover, miR‐3184 overexpression shortened the survival time of nude mice, while miR‐3184 silencing prolonged the life span (Figures 1N and S2M). To identify the molecular changes related to tumor promotion, we examined the expression of Ki67 and N‐cadherin in xenografted glioma samples. Ki67‐ and N‐cadherin‐positive cells were increased in the miR‐3184 overexpression group but decreased in the miR‐3184 silencing group, indicating a regulatory role for miR‐3184 in proliferation and epithelial‐mesenchymal transition (EMT) in glioma (Figure S2N). In the aforementioned studies, we confirmed the glioma‐promoting role of miR‐3184 in vitro and in vivo. To further study the molecules and pathways influenced by miR‐3184, we performed mRNA sequencing in miR‐3184‐overexpressing glioma cells. We found that overexpression of miR‐3184 was related to extracellular matrix organization, cell migration, positive regulation of cell proliferation, and the WNT signaling pathway (Figure 1O). Based on the analysis of GO enrichment, we performed western blot assays to verify the function of miR‐3184. We found that overexpression of miR‐3184 increased the expression of proteins related to migration, EMT, apoptosis, cell proliferation, and the Wnt‐β‐catenin pathway. However, silencing miR‐3184 induced the opposite effects on these factors (Figure 1P). In summary, these findings suggest that miR‐3184‐3p is enriched in pre‐CSF exosomes and promotes progression in gliomas.
FIGURE 1.
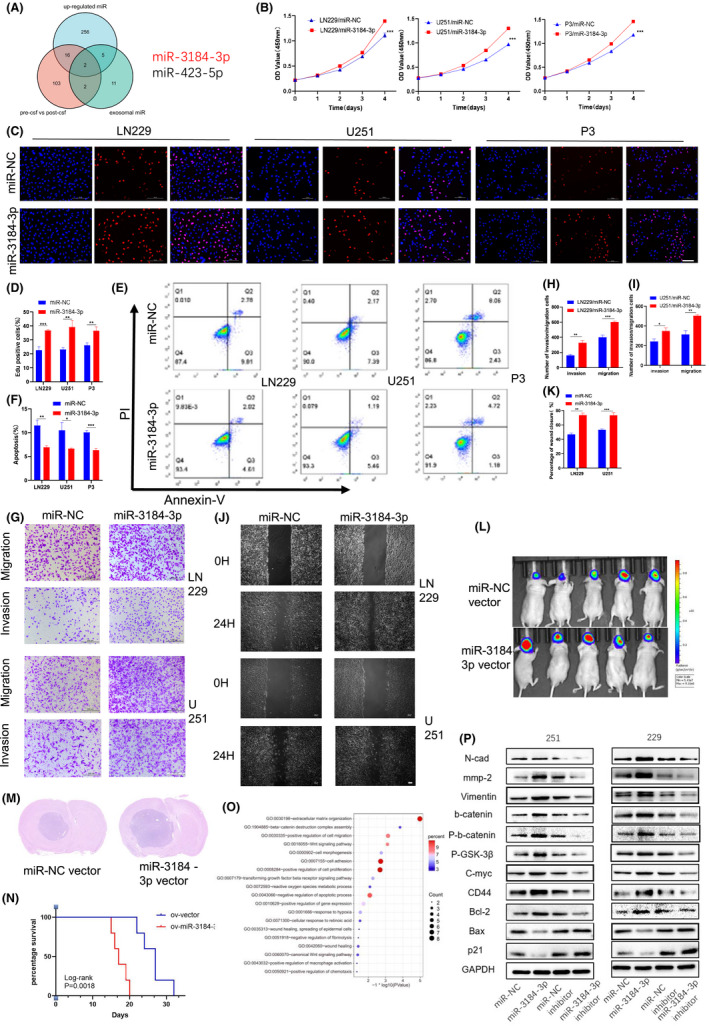
Pre‐CSF exosome enriched miR‐3184 promoted glioma progression. (A) Intersection of miRNA expression profile in patient samples. Exosomal miRNAs in CSF of glioma patients pre‐ (pre‐csf) and post‐ (post‐csf) tumor resection (red circle), exosomal miRNAs enriched in pre‐csf (green circle), and miRNAs upregulated in glioma tissue compared with normal brain tissue (blue circle) are shown. (B) The proliferation of glioma cells was examined by CCK8 assays after miR‐3184 overexpression. (C) The proliferation of glioma cells was examined by EdU assay after miR‐3184 overexpression. Scale bar = 100 µm. (D) Quantification of the results in (C). Data are presented as the means ± SD, n = 3 (*P < 0.05). (E) Apoptotic cells examined by flow cytometry. (F) Quantification of the results in (E). Data are presented as the means ± SD, n = 3 (*P < 0.05). (G) Invasion and migration abilities examined by Transwell assay. Scale bar = 200 µm. (H and I) Quantification of the results in (G). Data are presented as the means ± SD, n = 3 (*P < 0.05). (J) Wound healing assay examining glioma migration. Scale bar = 100 µm. (K) Quantification of the results in (J). Data are presented as the means ± SD, n = 3 (*P < 0.05). (L) Bioluminescence imaging showed the tumor sizes of each mouse, n = 10. (M) H&E staining of brain slices of mice in (L). (N) Kaplan–Meier survival curves for animals in each group. Log‐rank test, n = 10. (O) GO analysis for genes upregulated after miR‐3184 overexpression. (P) Western blot for proteins related to EMT, migration, proliferation, apoptosis, and wnt‐b‐catenin pathway [Correction added on 5 July 2022, after first online publication: in figure 1L, the image of the animal experiment data is wrong and is now corrected in the replacement figure].
3.2. PDCD4 is a direct target and mediates the protumor effect of miR‐3184
To identify the direct target of miR‐3184, we screened the downregulated mRNAs after miR‐3184 overexpression. We intersected these downregulated mRNAs with predicted miR‐3184 targets from two widely accepted prediction websites, TargetScan and miRDB (Figure 2A), and the drastically downregulated mRNA PDCD4 was chosen for further study (Figure 2B). We performed PCR and western blot assays to detect the changes in PDCD4 after miR‐3184 overexpression and inhibition. We found that both the mRNA and protein levels of PDCD4 decreased after miR‐3184 overexpression (Figure 2C,D). Additionally, we performed IHC staining for PDCD4 in xenografted glioma samples. PDCD4 expression was restored in the miR‐3184 silencing group, which confirmed our prediction (Figure S3A). A luciferase reporter assay was used to confirm the direct inhibition of PDCD4 by miR‐3184. The predicted binding site of miR‐3184 on the PDCD4 3′ untranslated region (UTR) is shown in Figure 2E. Luciferase activity was downregulated after miR‐3184 overexpression, indicating direct binding to these sequences (Figure 2F). We subsequently co‐overexpressed miR‐3184 and PDCD4 in glioma cells. The protumor effect of miR‐3184 was dramatically suppressed by PDCD4 overexpression. The increases in cell viability (Figure 2G), migration, and invasion (Figure 2J‐L) and the decrease in apoptosis (Figure 2H‐I) were offset by PDCD4 transfection, indicating that PDCD4 is a downstream target of miR‐3184 and mediates its protumor effect. The western blotting results were in accordance with the above results (Figure 2M). Furthermore, knockdown of PDCD4 in glioma cells mimicked the effects of miR‐3184 overexpression. After si‐PDCD4 transfection, glioma cells displayed enhanced proliferation, invasion, and migration abilities (Figure 3A–G). However, apoptosis was downregulated after PDCD4 inhibition (Figure 3H,I). The molecular changes after si‐PDCD4 transfection were similar to those after miR‐3184 overexpression (Figure 3J). These results demonstrate that PDCD4 is directly targeted by miR‐3184 and mediates the protumor function of miR‐3184.
FIGURE 2.
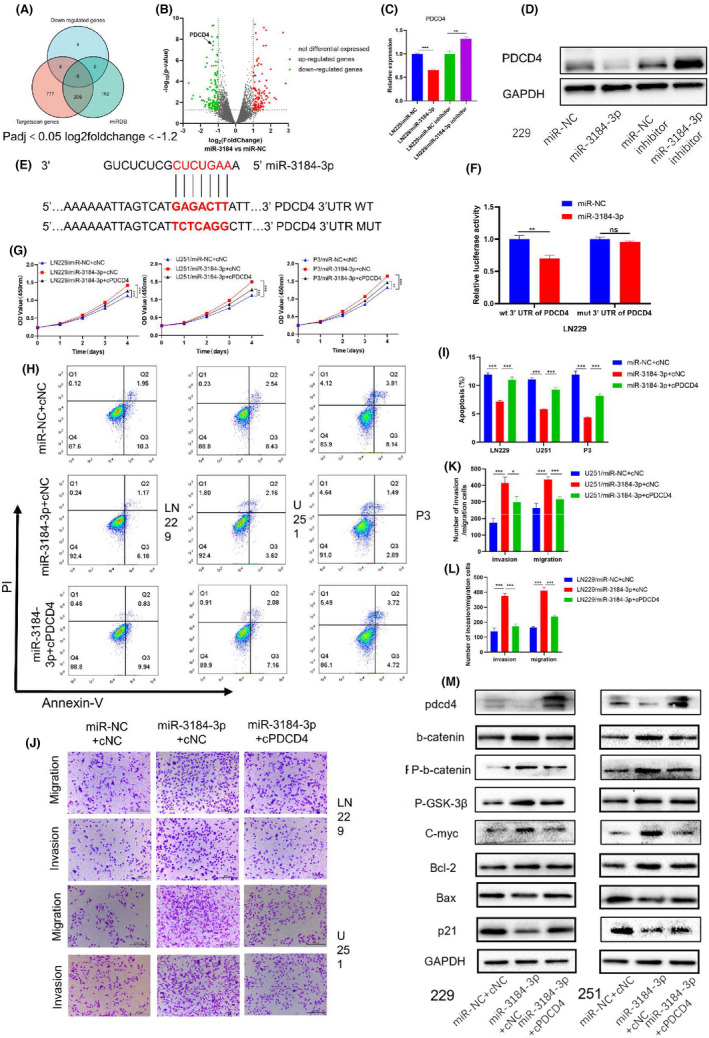
PDCD4 was a direct target of miR‐3184 and mediated the pro‐tumor effect. (A) Venn diagram showed the intersection of miR‐3184‐downregulated genes and predicting targets of miR‐3184 in two predicting websites. (B) Volcano plot showed the genes regulated by miR‐3184 overexpression. (C) PDCD4 mRNA expression after miR‐3184 overexpression or knockdown. Data are presented as the means ± SD, n = 3 (*P < 0.05). (D) PDCD4 mRNA expression after miR‐3184 overexpression or knockdown, n = 3. (E) Schematic representation of the predicted binding sites for miR‐3184 in the PDCD4 3′‐UTR (wild type, WT) and the designed mutant versions (mutant, MUT) of the PDCD4 3′‐UTR. (F) Relative luciferase activity of LN229 cells in the presence of the indicated treatments. Data are presented as the means ± SD, n = 3 (*P < 0.05). (G) The proliferation of glioma cells was examined by CCK8 assays. Data are presented as the means ± SD, n = 3 (*P < 0.05). (H) Apoptotic cells examined by flow cytometry. (I) Quantification of the results in (H). Data are presented as the means ± SD, n = 3 (*P < 0.05). (J) Invasion and migration abilities examined by Transwell assay. Scale bar = 200 µm. (K and L) Quantification of the results in (J). Data are presented as the means ± SD, n = 3 (*P < 0.05). (M) Western blot for PDCD4 and proteins related to EMT, migration, proliferation, apoptosis, and wnt‐b‐catenin pathway
FIGURE 3.
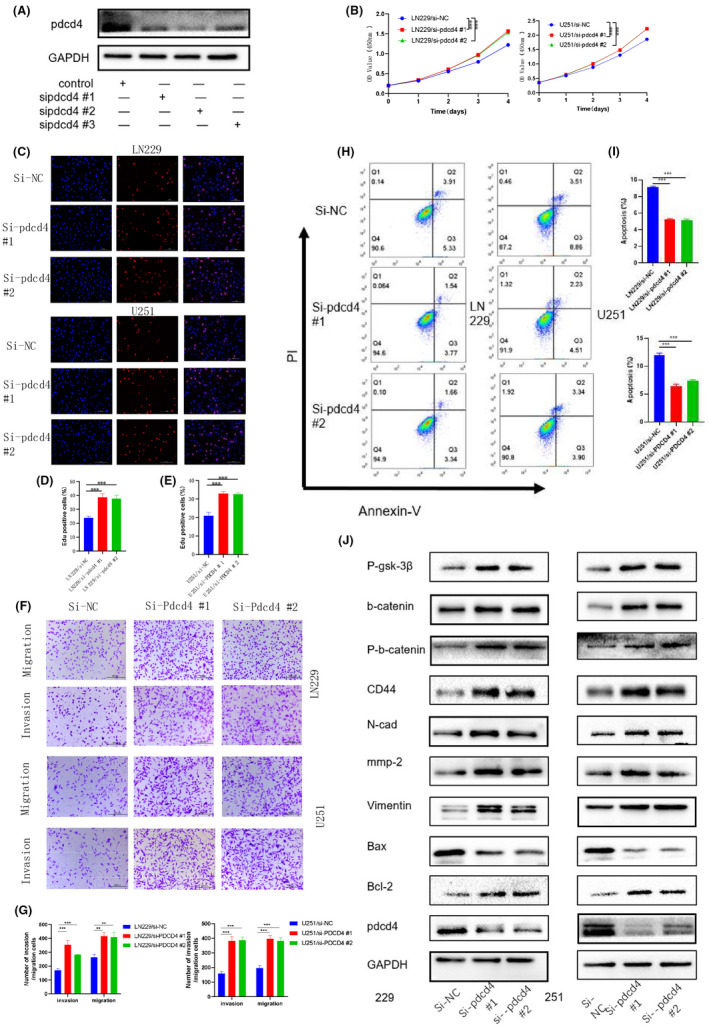
Silence PDCD4 displayed similar effects as miR‐3184 overexpression. (A) Western blot for PDCD4 after siRNA transfection, n = 3. (B) The proliferation of glioma cells was examined by CCK8 assays after PDCD4 knockdown. (C) The proliferation of glioma cells was examined by EdU assay. Scale bar = 100 µm. (D and E) Quantification of the results in (C). Data are presented as the means ± SD, n = 3 (*P < 0.05). (F) Invasion and migration abilities examined by Transwell assay. Scale bar = 200 µm. (G) Quantification of the results in (F). Data are presented as the means ± SD, n = 3 (*P < 0.05). (H) Apoptotic cells examined by flow cytometry. (I) Quantification of the results in (H). Data are presented as the means ± SD, n = 3 (*P < 0.05). (J) Western blot for PDCD4 and proteins related to EMT, migration, proliferation, apoptosis, and wnt‐b‐catenin pathway
3.3. Exosomal miR‐3184 induces macrophage M2‐like polarization
To confirm tumor cells secret exosomes containing miR‐3184‐3p, we collected exosomes from the supernatants of cultured GBM cells, which exhibited similar typical cup‐shaped morphology, size, and number (Figures 4F and S3B), and further confirmed their identity by detection of the exosome markers TSG101 and CD9 (Figure 4G), indicating that we successfully isolated exosomes from GBM cells. Furthermore, we collected exosomes from glioma cells transfected with miR‐NC, miR‐3184, sh‐miR‐NC, and sh‐miR‐3184. The miR‐3184 expression level was examined by PCR. We confirmed that the expression level of miR‐3184 in exosomes was concomitantly changed with cell transfection (Figure S3D). We also found that the expression of miR‐3184‐3p in exosomes was downregulated in cells treated by neutral sphingomyelinase‐2 (nSMase) GW4869, which blocks exosome formation (Figure S3E), thus confirming the existence of mir‐3184‐3p in exosomes. We wondered whether exosomal miR‐3184 could alter the immune microenvironment in glioma. We have previously reported that glioma cell‐derived exosomal miR‐1246 promotes macrophage M2‐like polarization. 6 We therefore examined the role of glioma cell exosomal miR‐3184 on macrophages. First, we tested the effects of miR‐3184 on macrophages. We found that after miR‐3184 transfection, both THP‐1‐induced macrophages and PBMC‐induced macrophages were polarized to an M2‐like phenotype, displaying an increased proportion of CD163‐positive cells (Figure 4A–D). In addition, miR‐3184 upregulated immunosuppressive mRNA expression in macrophages. The M1‐like molecule TNF‐a was concomitantly downregulated after miR‐3184 transfection in macrophages (Figure 4E). These results indicate that miR‐3184 could promote macrophage M2‐like polarization. Then, we collected exosomes from conditioned medium of LN229 and U251 glioma cell lines. Macrophages were cocultured with stained glioma‐derived exosomes and 24 h later fluorescence could be detected in macrophages, indicating the absorption of exosomes by macrophages (Figure S3C). We subsequently used these transfected exosomes for macrophage treatments. We found that sh‐miR‐NC vector exosomes could elevate CD163 expression in macrophages, whereas miR‐3184 silencing in exosomes partially blocked this effect. Exosomes with higher miR‐3184 expression showed stronger CD163 induction, which suggested that glioma‐derived exosomal miR‐3184 promotes the M2‐like polarization of macrophages (Figures 4H,I and S3F,G). In addition, other factors related to M2‐like polarization were regulated by exosomal miR‐3184 (Figure 4J). We further confirmed the abovementioned findings in vivo. According to our previous study, 6 we overexpressed miR‐3184 in macrophages and coinjected these cells with glioma cell lines into nude mice. We found that overexpression of miR‐3184 in macrophages significantly promoted tumor growth in vivo (Figure 4K,L) and shortened the survival times of mice (Figure 4M). The results of IHC staining also showed that miR‐3184 overexpression in macrophages increased the cell proliferation rate in xenografted glioma tissue (Figure 4N). Furthermore, we studied the role of macrophage transfected with miR 3184‐3p in the proliferation, invasion, and migration of glioma cells using the in vitro coculture system. The CCK8 and EDU assay results showed that glioma cells cocultured with miR‐3184‐3p overexpressing macrophages have better proliferation ability (Figure S4A,B). The invasion and migration assays demonstrated that macrophages transfected with miR‐3184‐3p promote glioma cell invasion and migration (Figure S4C,D). These results showed that glioma exosome‐derived miR‐3184‐3p contributed to M2 macrophages polarization. The M2 phenotype macrophages promoted the proliferation, invasion, and migration of glioma cells in vitro.
FIGURE 4.
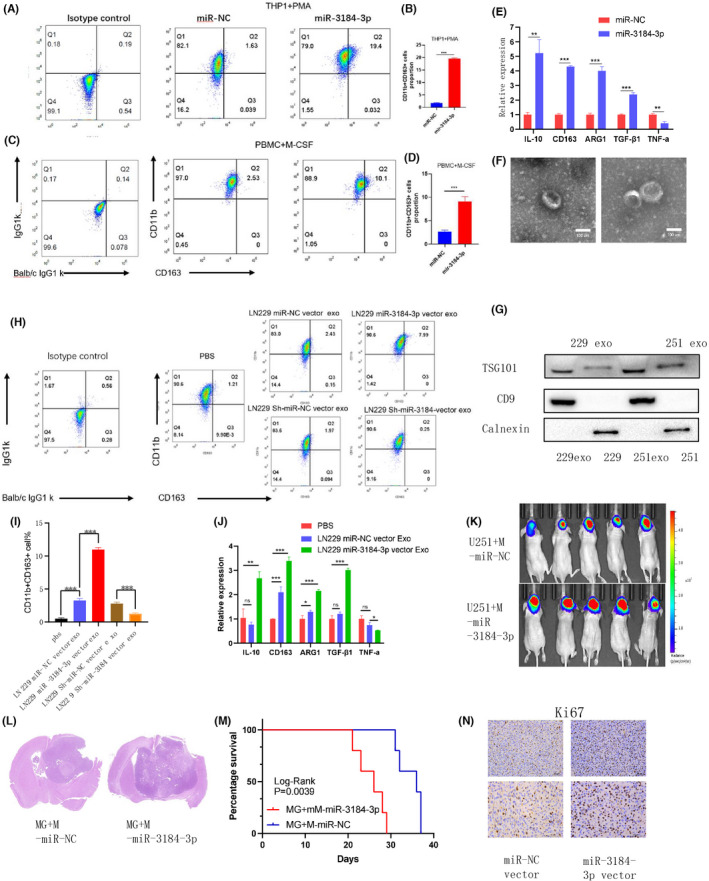
Exosomal miR‐3184 promoted M2‐like polarization in macrophages. (A) CD11b and CD163 positive cells examined by flow cytometry. (B) Quantification of the results in (A). Data are presented as the means ± SD, n = 3 (*P < 0.05). (C) CD11b and CD163 positive cells examined by flow cytometry. (D) Quantification of the results in (C). Data are presented as the means ± SD, n = 3. (E) mRNA expression of M1 and M2 markers after miR‐3184 overexpression in macrophages. Data are presented as the means ± SD, n = 3 (*P < 0.05). (F) Representative images for exosomes. Scale bar = 100 nm. (G) Western blot for exosome markers. (H) CD11b and CD163 positive cells examined by flow cytometry. (I) Quantification of the results in (H). Data are presented as the means ± SD, n = 3 (*P < 0.05). (J) mRNA expression of M1 and M2 markers in macrophages after different exosome treatments. Data are presented as the means ± SD, n = 3 (*P < 0.05). (K) Bioluminescence imaging showed the tumor sizes of each mouse, n = 10. (L) H&E staining of brain slices of mice in K. (M) Kaplan–Meier survival curves for animals in each group. Log‐rank test, n = 10. (N) Representative images of the IHC staining of Ki67, n = 3. Scale bar = 50 μm
3.4. miR‐3184 promotes macrophage M2‐like polarization via RSAD2
To identify the downstream target of miR‐3184, we performed mRNA sequencing for macrophages overexpressing miR‐3184. We then intersected the downregulated genes with predicted targets for miR‐3184 in the TargetScan website (Figure S4E). The gene RSAD2 was finally found (Figure 5A), of which the 3′ UTR contained potential binding sites for miR‐3184 (Figure 5B). We cotransfected RSAD2 and miR‐3184 in macrophages. Although RSAD2 transfection significantly increased the expression of RSAD2, cotransfection of RSAD2 and miR‐3184 attenuated this increase (Figure 5C). Then we performed a luciferase reporter assay to further confirm the direct binding of miR‐3184 with RSAD2 (Figure 5D). We confirmed that RSAD2 overexpression attenuated miR‐3184‐induced M2‐like polarization in macrophages via flow cytometry, ELISA, and PCR assays (Figure 5E–H). In addition, knockdown of RSAD2 in macrophages promoted the M2‐like phenotype, further suggesting that the role of miR‐3184 is mediated by RSAD2 (Figures 5I–L and S4F). It is widely accepted that the NF‐κB pathway plays an important role in the M2‐like polarization of macrophages, 17 thus we examined the protein levels of p65 and p‐p65 in miR‐3184‐altered cells. The results showed that overexpression of miR‐3184 or knockdown of RSAD2 inhibited the activation of p65, and p‐p65 expression was rescued when miR‐3184 and RSAD2 were co‐overexpressed in glioma cells (Figure 5M). Moreover, miR‐3184‐overexpressing glioma‐derived exosomes decreased the protein expression of RSAD2 and inhibited the phosphorylation of p65 (Figure 5N). In summary, we confirmed that glioma exosome miR‐3184 promotes macrophage polarization via RSAD2.
FIGURE 5.
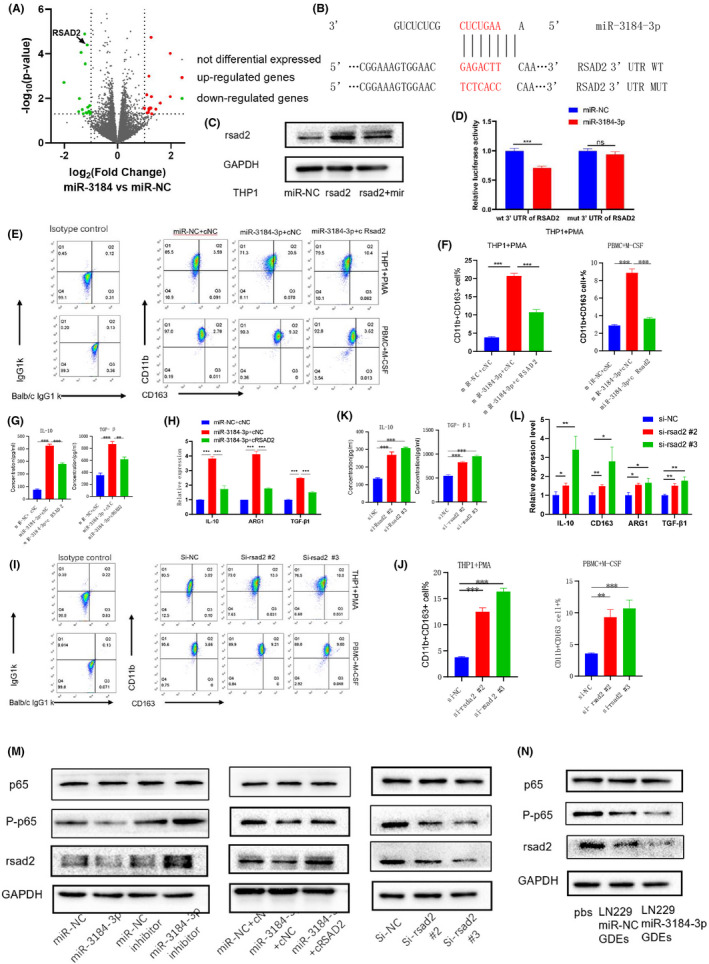
miR‐3184 promotes macrophage M2‐like polarization via RSAD2. (A) Volcano plot showed the genes regulated by miR‐3184 overexpression in macrophages. (B) Schematic representation of the predicted binding sites for miR‐3184 in the RSAD2 3′‐UTR (wild type, WT) and the designed mutant versions (mutant, MUT) of the RSAD2 3′‐UTR. (C) Western blot for RSAD2 in RSAD2‐ or RSAD2/miR‐3184‐double‐overexpressed macrophages. (D) Relative luciferase activity of macrophages in the presence of the indicated treatments. Data are presented as the means ± SD, n = 3 (*P < 0.05). (E) Apoptotic cells examined by flow cytometry. (F) Quantification of the results in (E). Data are presented as the means ± SD, n = 3 (*P < 0.05). (G) Cytokine secretion examined by ELISA. Data are presented as the means ± SD, n = 3 (*P < 0.05). (H) mRNA expression of M2 markers in macrophages after transfection. Data are presented as the means ± SD, n = 3 (*P < 0.05). (I) Apoptotic cells examined by flow cytometry. (J) Quantification of the results in (I). Data are presented as the means ± SD, n = 3 (*P < 0.05). (K) Cytokine secretion examined by ELISA. Data are presented as the means ± SD, n = 3 (*P < 0.05). (L) mRNA expression of M2 markers in macrophages after transfection. Data are presented as the means ± SD, n = 3 (*P < 0.05). (M) Western blot for p65, p‐p65, and RSAD2 after transfection in macrophages. (N) Western blot for p65, p‐p65, and RSAD2 after different exosomes treatment in macrophages
4. DISCUSSION
In the current study, we revealed the glioma‐promoting role of miR‐3184. On the one hand, miR‐3184 enhances glioma tumorigenesis by targeting PDCD4. On the other hand, miR‐3184 is enriched in glioma‐derived exosomes and induces M2‐like polarization in macrophages, which in turn aggravates glioma malignancy (Figure 6). These findings added new evidence that the interaction between glioma and the tumor‐associated immunosuppressive microenvironment facilitates glioma progression.
FIGURE 6.
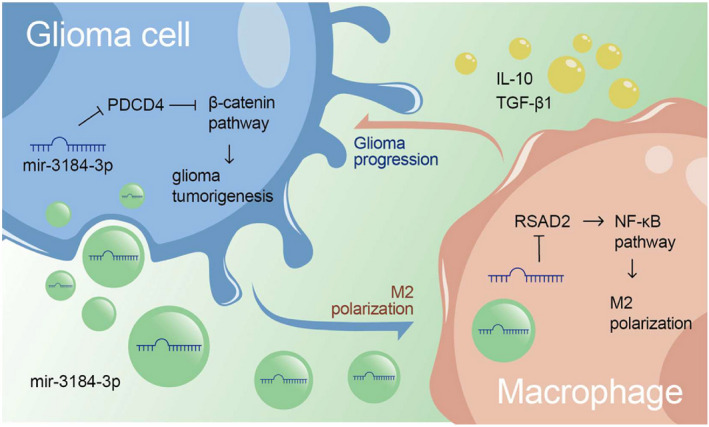
Illustration of the conclusions: miR‐3184‐3p promoted glioma progression in two ways. On the one hand, miR‐3184 enhanced glioma tumorigenesis via targeting PDCD4. On the other hand, miR‐3184 was enriched in glioma‐derived exosomes and induced M2‐like polarization in macrophages, which in return aggravated the glioma malignancy
Glioma remains an incurable disease with extremely poor outcomes. However, the sensitivity of current diagnostic tools such as magnetic resonance imaging (MRI) are not satisfactroy. 18 Early diagnosis and timely monitoring of therapeutic effectiveness are urgently needed. Recently, it was reported that liquid biopsy enables the noninvasive detection of central nervous system (CNS) tumors. For example, miRNAs in exosomes derived from body fluid are reflective of the cell of origin. 19 , 20 Hence, it is possible to use exosomal miRNAs as diagnostic factors for glioma. Considering the complex origin of plasma exosomes, 21 we screened the miRNA profile of CSF exosomes instead. We found a significant enrichment of miR‐3184 in exosomes from the CSF of preoperative patients and after tumor resection its expression level decreased concomitantly. Therefore, we inferred that the downregulation of miR‐3184 in CSF exosomes is a consequence of decreased tumor burden. Accordingly, further miRNA sequencing of glioma tissue showed that miR‐3184 is a glioma‐enriched miRNA, which confirmed our hypothesis that upregulated miR‐3184 in glioma could be secreted into exosomes and released into CSF.
MiR‐3184 is a recently defined miRNA regulating tumor malignancy. 22 In the current study, we first found that miR‐3184 promotes the progression of glioma cells. These protumor effects were mediated by the targeting of PDCD4. Furthermore, we proved that miR‐3184 could be transferred to macrophages via exosomes and induce M2‐like polarization of macrophages by targeting RSAD2.
Programmed cell death 4 (PDCD4) is a tumor suppressor whose expression is frequently downregulated in various types of cancers. PDCD4 functions as a protein translation inhibitor in tumors and thereby blocks pathways involving cell proliferation, survival, and invasion. 23 In the current study, we confirmed the antitumor role of PDCD4 in glioma. We found that knockdown of PDCD4 induced the activation of factors in the Wnt‐b‐catenin pathway and EMT process. These results were in accordance with previous findings that PDCD4 is a suppressor of EMT in tumors. 24 , 25 , 26
In Figure 5A, when overexpressing mir‐3184‐3p, we found the changes of many genes related to angiogenesis and immunosuppression. 27 We determined that Radical S‐adenosylmethionine (SAM) domain‐containing protein 2 (RSAD2) downregulated is one of the most significant genes and may be the directly downstream targets of miR‐3184‐3p. RSAD2 is a key enzyme in innate immune responses that is highly expressed in response to viral infection and inflammatory stimuli in many cell types. 28 Among human THP‐1‐induced macrophages, RSAD2 is upregulated in M1 subtype macrophages, indicating a correlation between RSAD2 and M1‐like polarization. 29 Our study found that inhibition of RSAD2 by miR‐3184 in macrophages induced M2‐like polarization.
In summary, we uncovered a novel interaction between glioma cells and TAMs that enhances tumor progression. miR‐3184, which functions as a key regulator during this process, is probably a predictive factor for glioma progression. However, there are some limitations to this study. The diagnostic ability of miR‐3184 should be further assessed, and the detailed downstream pathway by which miR‐3184‐PDCD4 or miR‐3184‐RSAD2 induces tumor progression needs further study.
DISCLOSURE
The authors declare that they have no competing interests.
AUTHOR CONTRIBUTIONS
G.L. supervised the project. H.X. designed the research and performed all experiments. M.L. and Z.W.P. completed the basic experimental part. Z.P.Z., Z.J.G., and R.R.Z. performed statistical analysis with the R language. B.Y.L., Y.H.Q., Q.D.G., S.J.Z., S.L.Z., and S.B.W. were responsible for clinical sample collection and subsequent sample delivery. X.G., L.D., and H.X. helped to revise the manuscript. All authors read and approved the final manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The research protocol was reviewed and approved by the Ethical Committee on Scientific Research of Shandong University Qilu Hospital (approval number: KYLL‐2018‐324), and written informed consent was obtained from each patient included in the study. The patient data were acquired from publicly available datasets, which contained complete informed consent information for the patients.
ANIMAL EXPERIMENTS
All procedures that involved mice were approved by the Animal Care and Use Committee of the Qilu Hospital of Shandong University.
Supporting information
Figure S1‐4
Table S1
Table S2
Method S1
ACKNOWLEDGMENTS
We thank the surgeons and patients who participated in these studies, and Novogene Co. Ltd and Tianjin Novogene Bioinformatics Technology Co. Ltd for miRNA sequencing and mRNA sequencing technical development and support.
Xu H, Li M, Pan Z, et al. miR‐3184‐3p enriched in cerebrospinal fluid exosomes contributes to progression of glioma and promotes M2‐like macrophage polarization. Cancer Sci. 2022;113:2668–2680. doi: 10.1111/cas.15372
Funding information
This work was supported by grants from the National Natural Science Foundation of China (Nos. 81874083, 82072776, 82072775, 81702468, 81802966, 81902540, 81874082, 81472353), the Natural Science Foundation of Shandong Province of China (Nos. ZR2019BH057, ZR2020QH174, ZR2021LSW025), the Jinan Science and Technology Bureau of Shandong Province (2021GXRC029), the Key Clinical Research Project of the Clinical Research Center of Shandong University (2020SDUCRCA011) and the Taishan Pandeng Scholar Program of Shandong Province (No. tspd20210322)
DATA AVAILABILITY STATEMENT
All data used in this work can be acquired from the TCGA database (http://cancergenome.nih.gov/) and the CGGA database (http://www.cgga.org.cn/), and the miRNA sequencing and mRNA sequencing data of our local samples have been deposited in the Genome Sequence Archive (GSA) under accession number CRA002339. The data were released when the paper was published. The processed data are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Wen P, Reardon DA. Neuro‐oncology in 2015: progress in glioma diagnosis, classification and treatment. Nat Rev Neurol. 2016;12(2):69‐70. [DOI] [PubMed] [Google Scholar]
- 2. Thakkar J, Dolecek T, Horbinski C, et al. Epidemiologic and molecular prognostic review of glioblastoma. Cancer Epidemiol Biomark Prev. 2014;23(10):1985‐1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lombardi G, Caccese M, Padovan M, et al. Regorafenib in recurrent glioblastoma patients: a large and monocentric real‐life study. Cancers. 2021;13(18):4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Charles N, Holland E, Gilbertson R, Glass R, Kettenmann HJG. The brain tumor microenvironment. Glia. 2011;59(8):1169‐1180. [DOI] [PubMed] [Google Scholar]
- 5. Guo X, Xue H, Shao Q, et al. Hypoxia promotes glioma‐associated macrophage infiltration via periostin and subsequent M2 polarization by upregulating TGF‐beta and M‐CSFR. Oncotarget. 2016;7(49):80521‐80542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Qian M, Wang S, Guo X, et al. Hypoxic glioma‐derived exosomes deliver microRNA‐1246 to induce M2 macrophage polarization by targeting TERF2IP via the STAT3 and NF‐κB pathways. Oncogene. 2020;39(2):428‐442. [DOI] [PubMed] [Google Scholar]
- 7. Zhang Z, Xu J, Chen Z, et al. Transfer of MicroRNA via macrophage‐derived extracellular vesicles promotes proneural‐to‐mesenchymal transition in glioma stem cells. Cancer Immunol Res. 2020;8(7):966‐981. [DOI] [PubMed] [Google Scholar]
- 8. Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2(8):569‐579. [DOI] [PubMed] [Google Scholar]
- 9. Kalluri R, LeBleu VJS. The biology function and biomedical applications of exosomes. Science. 2020;367:eaau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jones J, Nguyen H, Drummond K, Morokoff AJN. Circulating biomarkers for glioma: a review. Neurosurgery. 2021;88(3):E221‐E230. [DOI] [PubMed] [Google Scholar]
- 11. Kong Y, Ferland‐McCollough D, Jackson TJ, Bushell M. microRNAs in cancer management. Lancet Oncol. 2012;13(6):e249‐e258. [DOI] [PubMed] [Google Scholar]
- 12. Xu S, Zhang J, Xue H, et al. MicroRNA‐584‐3p reduces the vasculogenic mimicry of human glioma cells by regulating hypoxia‐induced ROCK1 dependent stress fiber formation. Neoplasma. 2017;64(1):13‐21. [DOI] [PubMed] [Google Scholar]
- 13. Zhang Z, Guo X, Guo X, et al. MicroRNA‐29a‐3p delivery via exosomes derived from engineered human mesenchymal stem cells exerts tumour suppressive effects by inhibiting migration and vasculogenic mimicry in glioma. Aging. 2021;13(4):5055‐5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jafari D, Tiyuri A, Rezaei E, et al. Diagnostic accuracy of cerebrospinal fluid and serum‐isolated extracellular vesicles for glioblastoma: a systematic review and meta‐analysis. Expert Rev Mol Diagn. 2020;20(11):1075‐1085. [DOI] [PubMed] [Google Scholar]
- 15. Guo X, Qiu W, Wang J, et al. Glioma exosomes mediate the expansion and function of myeloid‐derived suppressor cells through microRNA‐29a/Hbp1 and microRNA‐92a/Prkar1a pathways. Int J Cancer. 2019;144(12):3111‐3126. [DOI] [PubMed] [Google Scholar]
- 16. Bang C, Batkai S, Dangwal S, et al. Cardiac fibroblast–derived microRNA passenger strand‐enriched exosomes mediate cardiomyocyte hypertrophy. J Clin Invest. 2014;124(5):2136‐2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li H, Jiang T, Li M, Zheng X, Zhao G‐J. Transcriptional regulation of macrophages polarization by microRNAs. Front Immunol. 2018;9:1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grossman S, Batara JF. Current management of glioblastoma multiforme. Semin Oncol. 2004;31(5):635‐644. [DOI] [PubMed] [Google Scholar]
- 19. Akers J, Ramakrishnan V, Kim R, et al. miR‐21 in the extracellular vesicles (EVs) of cerebrospinal fluid (CSF): a platform for glioblastoma biomarker development. PLoS One. 2013;8(10):e78115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Santiago‐Dieppa D, Steinberg J, Gonda D, Cheung V, Carter B, Chen CC. Extracellular vesicles as a platform for ‘liquid biopsy’ in glioblastoma patients. Expert Rev Mol Diagn. 2014;14(7):819‐825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Notarangelo M, Zucal C, Modelska A, et al. Ultrasensitive detection of cancer biomarkers by nickel‐based isolation of polydisperse extracellular vesicles from blood. EBioMedicine. 2019;43:114‐126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lin D, Fu Z, Yang G, et al. Exportin‐5 SUMOylation promotes hepatocellular carcinoma progression. Exp Cell Res. 2020;395(2):112219. [DOI] [PubMed] [Google Scholar]
- 23. Wang Q, Yang H‐S. The role of Pdcd4 in tumour suppression and protein translation. Biol Cell. 2018;110(8):169‐177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang Q, Sun Z, Yang HJO. Downregulation of tumor suppressor Pdcd4 promotes invasion and activates both beta‐catenin/Tcf and AP‐1‐dependent transcription in colon carcinoma cells. Oncogene. 2008;27(11):1527‐1535. [DOI] [PubMed] [Google Scholar]
- 25. Wang Q, Sun Z, Allgayer H, Yang HJO. Downregulation of E‐cadherin is an essential event in activating beta‐catenin/Tcf‐dependent transcription and expression of its target genes in Pdcd4 knockdown cells. Oncogene. 2010;29(1):128‐138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang Q, Zhu J, Zhang Y, et al. Down‐regulation of programmed cell death 4 leads to epithelial to mesenchymal transition and promotes metastasis in mice. Eur J Cancer. 2013;49(7):1761‐1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Komohara Y, Jinushi M, Takeya M. Clinical significance of macrophage heterogeneity in human malignant tumors. Cancer Sci. 2014;105(1):1‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Honarmand Ebrahimi K, Vowles J, Browne C, McCullagh J, James WS. ddhCTP produced by the radical‐SAM activity of RSAD2 (viperin) inhibits the NAD ‐dependent activity of enzymes to modulate metabolism. FEBS Lett. 2020;594(10):1631‐1644. [DOI] [PubMed] [Google Scholar]
- 29. Li P, Hao Z, Wu J, et al. Comparative proteomic analysis of polarized human THP‐1 and mouse RAW264.7 macrophages. Front Immunol. 2021;12:700009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1‐4
Table S1
Table S2
Method S1
Data Availability Statement
All data used in this work can be acquired from the TCGA database (http://cancergenome.nih.gov/) and the CGGA database (http://www.cgga.org.cn/), and the miRNA sequencing and mRNA sequencing data of our local samples have been deposited in the Genome Sequence Archive (GSA) under accession number CRA002339. The data were released when the paper was published. The processed data are available from the corresponding author upon reasonable request.


