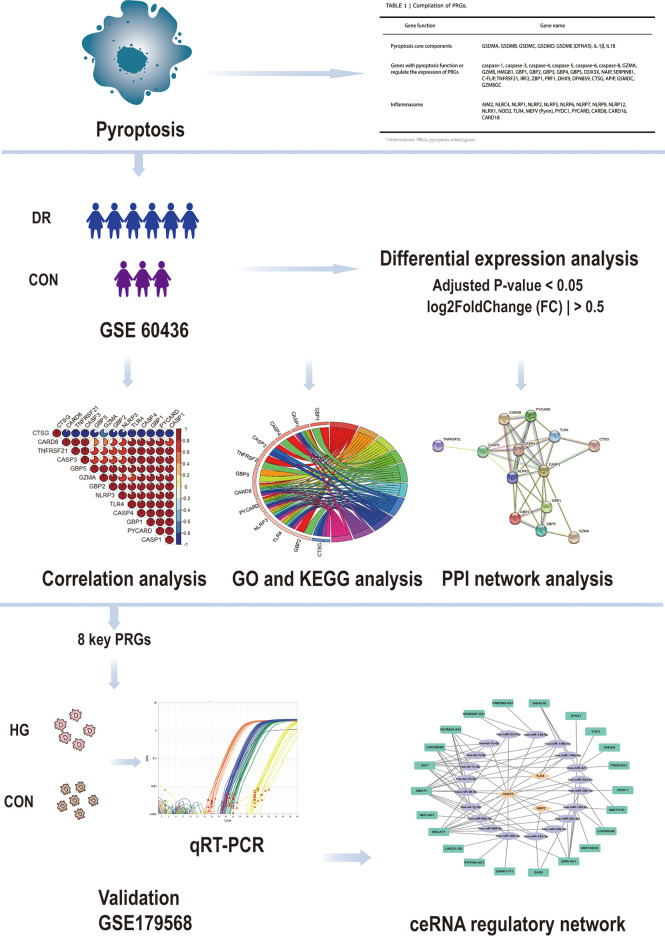
Figure 1.
Design idea of this study. Downloaded GSE60436 dataset from the GEO database and 51 PRGs collected from the PubMed database. The R software was used to process the data, such as quality control, normalization, and background correction. A total of 13 differentially expressed PRGs in the dataset were identified by difference analysis, and GO and KEGG enrichment analyses were performed. At the same time, eight hub genes were identified by PPI analysis and then verified with qRT-PCR experiments and GSE179568. Finally, the specific mechanism of PRGs in DR was revealed by structuring a ceRNA regulatory network through multiMiR database and starbase database. GEO, Gene Expression Omnibus; PRGs, pyroptosis-related genes; PPI, protein–protein interaction; DR, diabetic retinopathy; qRT-PCR, quantitative real-time polymerase chain reaction; ceRNA, competing endogenous RNAs; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.