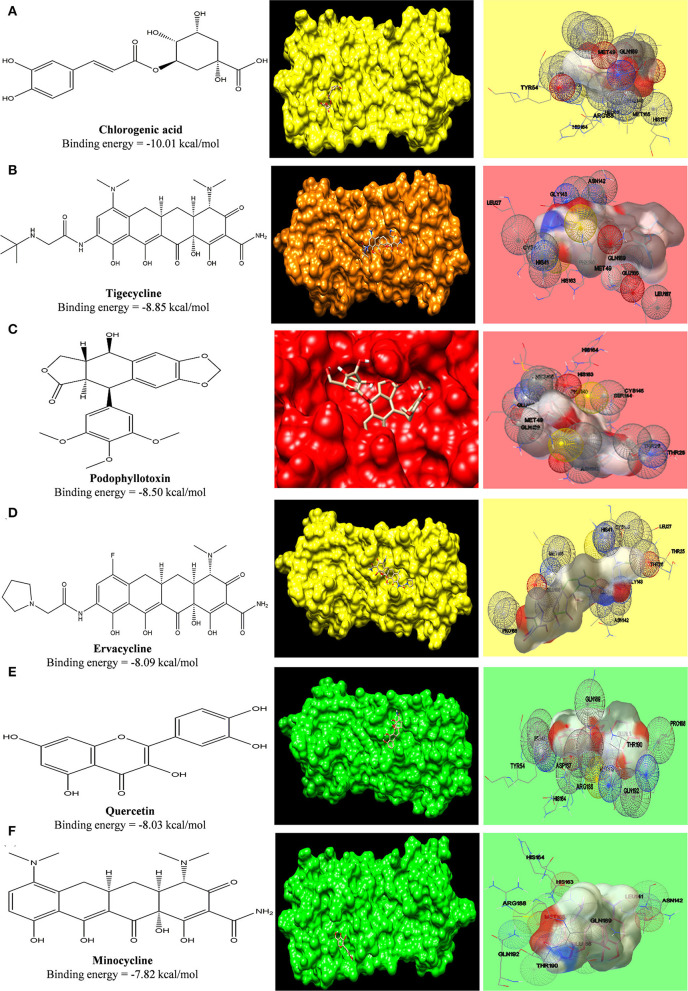
Figure 10.
Computed structural comparison and binding features (visualization using UCSF Chimera and AutoDock) of Chlorogenic acid (A), Tigecycline (B), Podophyllotoxin (C), Ervacycline (D), Quercetin (E), and Minocycline (F), with SARS-COV2 main protease. The lower the binding energy, the greater will be the binding affinity.