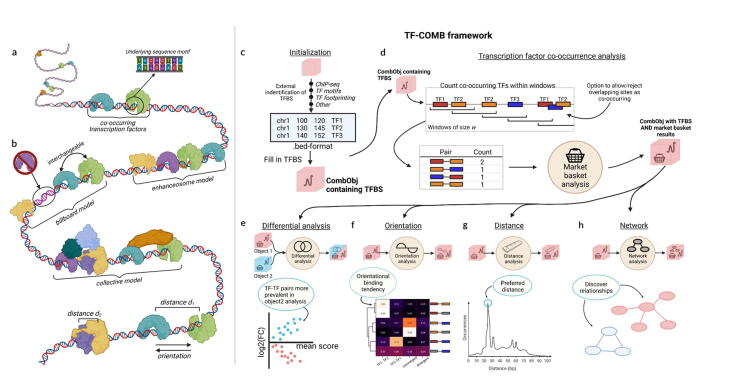
Fig. 1.
Grammar of regulatory elements and the TF-COMB framework. a) Concept of two co-occurring transcription factors (green + blue) bound in immediate proximity directly to the DNA. b) Models of TF-enhancer interactions and TF binding characteristics. The enhanceosome is defined by strict positioning of TFs, whereas the billboard allows for interchanged positions (green + blue) and absent TF factors (purple). The collective model allows TFs to bind on top of other factors (dark orange, light blue, dark green). TF pairs also exhibit additional characteristics, such as preferred binding distance (d1 and d2), as well as binding in different orientations on the DNA. Drawn with inspiration from [2]. c-h) The TF-COMB framework: c) Initialization of a TF-COMB object (red square) by providing TFBS and regions of interest from any data origin (e.g. ChIP-Seq, footprinting, ATAC-peaks). d) Co-occurrences are identified, counted and analyzed with an adapted market basket analysis, and are stored in the object for further analyses. e) Differential analysis module allows for the comparison of two independent TF-COMB objects. The module visualizes data to indicate TF pairs more frequent in Object1 (red dots) and Object2 (blue dots) respectively. f) The orientational binding module calculates strand specificity of TF pairs and visualizes preferences via heatmaps. g) TF pairs are analyzed in context of their binding distance, and pairs with prefered binding distance are classified and visualized as histograms. h) Network analysis and visualization module allows to identify higher order relationships between TFs and/or other features. All subfigures were created with BioRender.com. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)