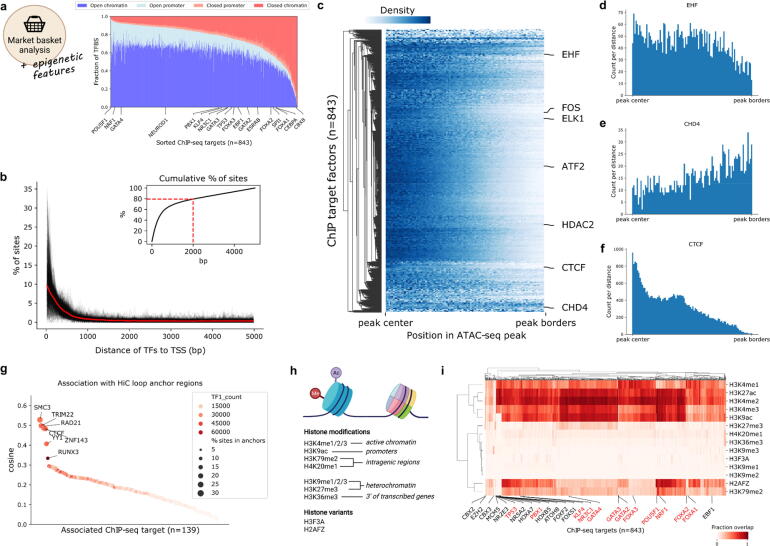
Fig. 3.
Integration of epigenetic marks reveals positional identity of TFs and co-factors. a) Percentage of ChIP-seq peaks (y-axis) in open/closed (defined by overlap with ATAC-seq peaks) promoter regions (defined as 5000 bp upstream of the TSS) and chromatin regions (all regions not defined as promoter) for individual factors (x-axis). b) Distance of ChIP-seq peak summits to transcription start sites (TSS) of genes. Distributions for individual factors are shown in black, and the mean distance is shown in bold red. Upper right corner shows the cumulative distribution of sites with 80% of sites marked with a dashed line. c) Relative location of ChIP-seq peak summits in open chromatin regions. Counts for the same TF in different cell lines were merged by taking the mean at each distance. The colorbar represents the scaled number of positions found to co-localize at different percentages of the peak length. Counts to the left/right of the peak center are aggregated to a range from center to border. d-f) Relative TF binding positions in ATAC-seq peaks from peak-center (left) to outer peak-borders (right). g) Co-occurrence of TFs and HiC loop regions in the cell line GM12878. TF1_count represents the total count of ChIP-seq peaks per factor. h) Scheme of histone modifications and histone variants investigated. Created with BioRender.com. i) Co-occurrence of TFs and histone marks as calculated by fractional overlap of regions. A selection of TFs from interesting clusters are annotated on the x-axis. Known pioneer factors are marked in red. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)