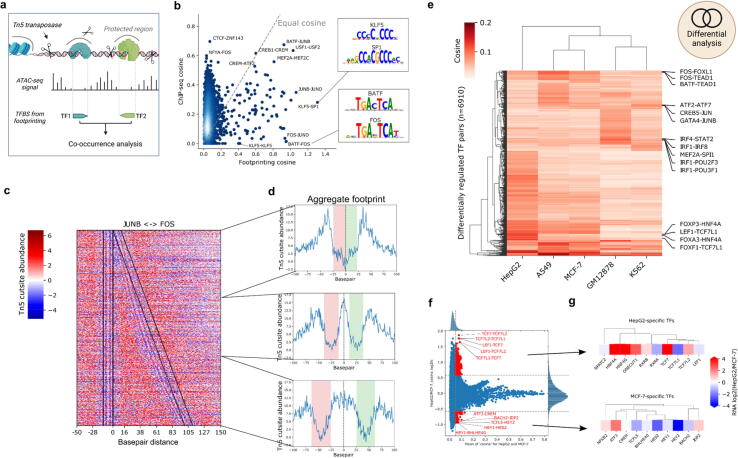
Fig. 4.
Footprinting data uncovers cell line specific TF co-occurrence. a) Scheme of TF co-occurrence and Tn5 mediated digital genomic footprinting. Prepared using BioRender.com. b) Direct comparison of scores derived for TF-pairs via ChIP-seq and ATAC-seq (footprinting) analysis. Two pairs with high footprinting scores are highlighted and corresponding motifs are illustrated. c) Footprinting heatmap of all co-occurring JUNB-FOS sites. Colored for Tn5 cutsites appearing more than expected (red) or less than expected (blue) if DNA is inaccessible. Black lines represent the edges of JUNB (left) and FOS (right) motifs. Edges show the binding strand of the respective TF. d) Aggregated views of the scores shown in e). Increasing distance (top to bottom) causes the combined TFs footprint to split into two distinct ones. e) Heatmap showing cosine scores for differentially co-occurring TF pairs across five cell lines. A subset of prominent cell line specific pairs are labeled on the right side. f) Activity of TF-pairs in direct comparison between HepG2 and MCF-7 cell lines. Significantly changed TF pairs are marked in red. g) Differential RNA expressions of the top 10 TFs selected in f) for each group. TFs are clustered by motif similarity. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)