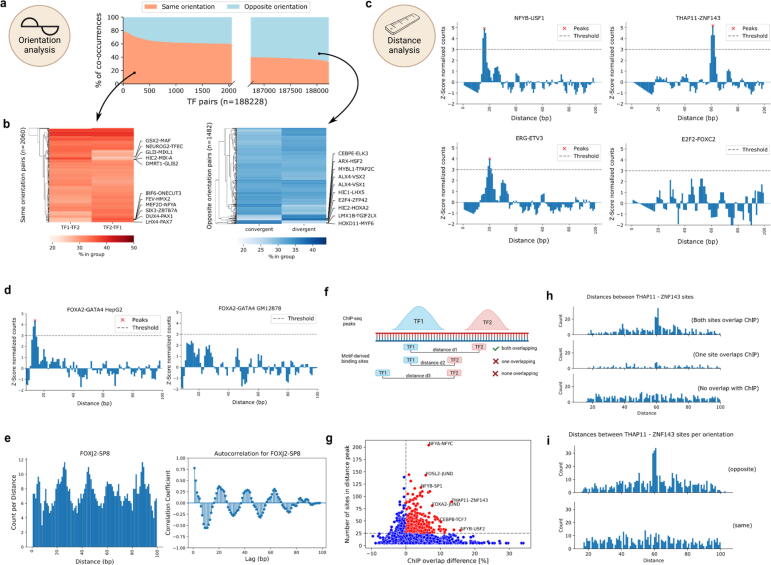
Fig. 5.
TF pairs exhibit local binding grammar. a) Percentage of TF-pair locations (y-axis) with both TFs on the same or opposite strand. X-axis gives the ranking of TF pairs with regards to orientation. Only pairs with more than 60% for either group are shown (axis is not continuous). b) Percentage of TF-pair locations derived from a), splitted by TFs orientation. Red heatmap highlights top pairs with orientation on the same strand, blue heatmap highlights top pairs with orientation on opposite strands. c) Z-score normalized TF-pair binding counts sorted by distance. Peaks above threshold are called by TF-COMB and considered preferred binding distance. d) Difference in binding distance distribution for FOXA2-GATA4 in HepG2 (left) and GM12878 (right) cells. e) Binding distance periodicity of the FOXJ2-SP8 pair. Left plot shows the distribution of binding site distances. Right plot shows the calculated autocorrelation for the signal, indicating a lag of 20 bp. f) Scheme of motif-derived binding sites overlapping with ChIP peaks. Prepared using BioRender.com. g) Difference in ChIP-overlap fraction (x-axis) between preferred distance sites and no preferred distance sites per pair and power (y-axis). The most prominent pairs are annotated. h) Number of sites by distance between THAP11 and ZNF143 binding sites. Subplots indicate split of loci into groups as indicated in f). i) Number of sites by distance between THAP11 and ZNF143 binding sites. Upper and lower plots indicate strand orientation. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)