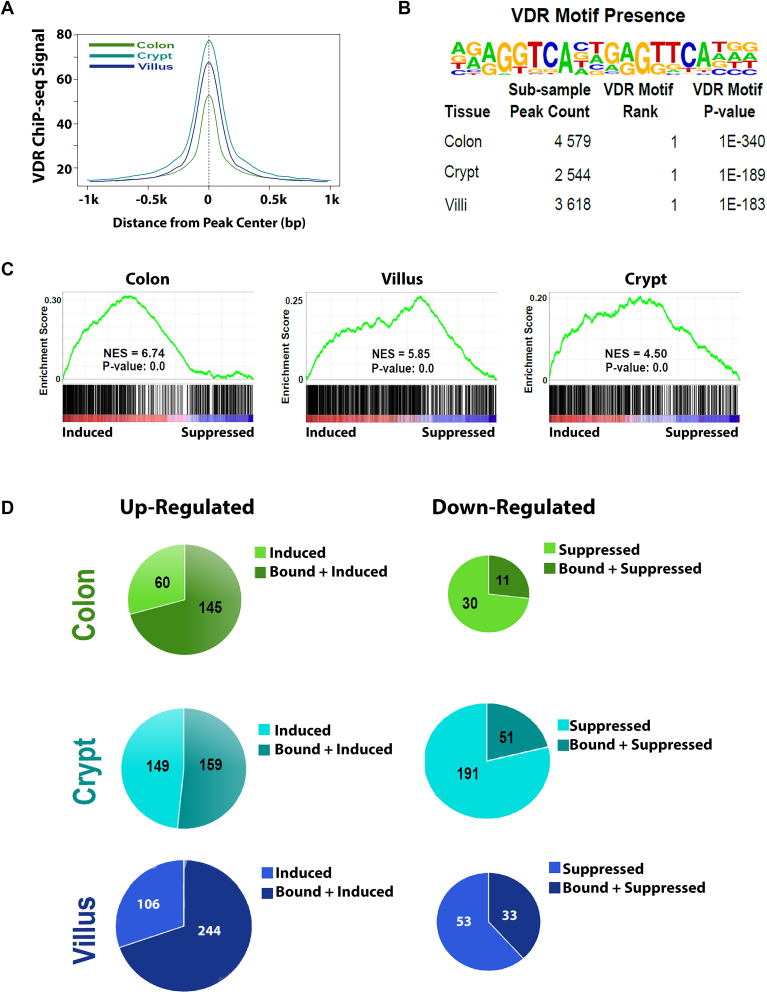
Figure 3.
ChIP-seq analysis of VDR binding and its relation to differentially expressed genes in each compartment. Mice were treated with 1,25(OH)2D3 (10 ng/g body weight) and tissues were collected for VDR ChIP-seq after 1 h. A, composite ChIP-seq signal at regions identified as binding VDR in the three intestinal compartments show strong and centered sequence reads. B, HOMER motif analysis identified transcription factor–binding motifs enriched at VDR-binding regions from each tissue. Twenty percent of the ChIP peaks identified in each tissue were randomly sampled for motif analysis. The canonical VDR motif was the most prevalent in each tissue. C, RNA-seq analysis was conducted in each tissue and genes linked to colon, crypt, or villus binding (within 10 kb) were correlated utilizing GSEA analysis. GSEA plots indicate VDR-binding sites are nearby genes that are 1,25(OH)2D3-induced in each tissue. D, pie charts depict genes upregulated (Left) or downregulated (Right) by 1,25(OH)2D3 treatment (5% FDR) and the subset of these bound by VDR in the colon, small intestinal crypt, or small intestinal villus. 1,25(OH)2D3, 1,25-dihydroxyvitamin D3; VDR, vitamin D receptor.