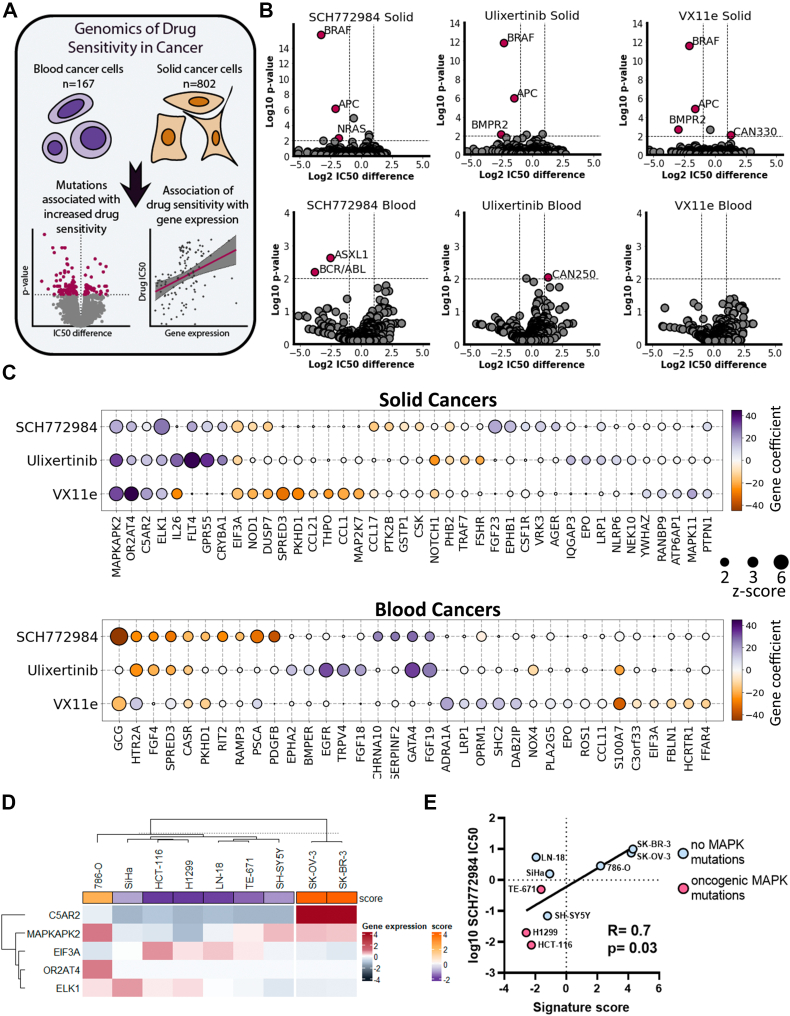
Figure 7.
Genetic and transcriptomic markers for sensitivity to ERK inhibitors.A, scheme for analysis design. B, volcano plots showing mutations associated with sensitivity/resistance in solid and blood cancers. Mutations with p-values <0.01 after FDR correction and IC50 change >2-fold were selected as significant. C, genes whose expression is associated with sensitivity/resistance as revealed by Elastic Net analysis. Gene coefficients correspond to what predicted impact gene expression has on IC50 values in linear regression model after elastic net regularization. Positive gene coefficients mean that gene expression increases drug IC50 and negative, decreases. Genes with z-scores >2 for at least one inhibitor are shown. D, K-means clustering of nine cancer cell lines based on expression of C5AR2, MAPKAPK2, EIF3A, OR2AT4, and ELK1 genes. Gene expression is shown as log2 difference in gene expression for a cell line relative to mean expression for all cell lines. Signature score (score) was calculated based on relative gene expression and Elastic Net regression results. E, Pearson correlation of log10 IC50 values for SCH772984 and signature scores. GDSC, Genomic of Drug Sensitivity in Cancer database.