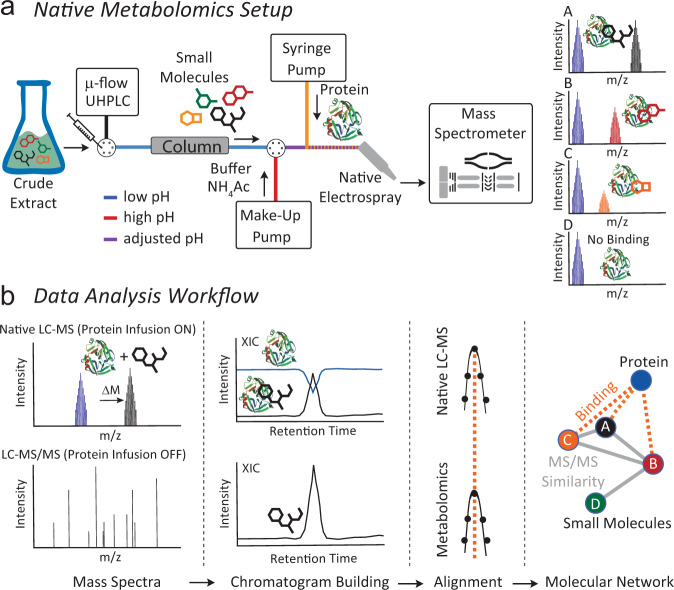
Fig. 1. Overview of the native metabolomics workflow.
a Native metabolomics setup. A crude extract is separated by µ-flow UHPLC. The pH is adjusted after chromatography with ammonium acetate to native-like conditions via the make-up pump. Orthogonally, the protein of interest is infused, and the resulting protein-binder complexes are measured by MS. The procedure is repeated as a metabolomics run (high-resolution UHPLC-MS/MS acquisition without protein infusion). For the data analysis (b) the Δ m/z and retention time of the native MS run are correlated with m/z values and retention time of the metabolomics (LC-MS/MS) run and subsequently visualized using molecular networking and retention time pairing that links the observed mass differences of the protein in the native state vs bound states with the parent mass of MS/MS spectra of the small molecule.