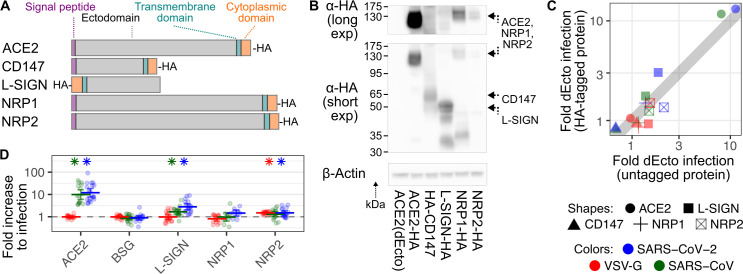
Fig 2. Established and proposed SARS-CoV-2 receptor proteins.
(A) Schematic showing relative lengths, tag locations, and overall domain topologies of proteins. (B) Representative immunoblot of cells stably expressing HA-tagged versions of each protein, including longer (top) and shorter (bottom) anti-HA exposures. Beta actin was blotted as a loading control. (C) Scatter plot comparing normalized pseudovirus infection rates of cells stably expressing various HA-tagged and untagged proteins. Shapes correspond to receptors. Colors correspond to viruses. (D) Compiled normalized infection data for the established and proposed SARS-CoV-2 receptor proteins. Error bars denote 95% confidence intervals for at least 10 replicates. Asterisks denote samples with p < 0.01. The underlying data can be found in S2 Data, and the source code can be found at https://github.com/MatreyekLab/ACE2_dependence. ACE2, angiotensin converting enzyme-2; SARS-CoV, Severe Acute Respiratory Syndrome-related Coronavirus; SARS-CoV-2, Severe Acute Respiratory Syndrome-related Coronavirus 2; VSV-G, vesicular stomatitis virus glycoprotein.