Figure 2.
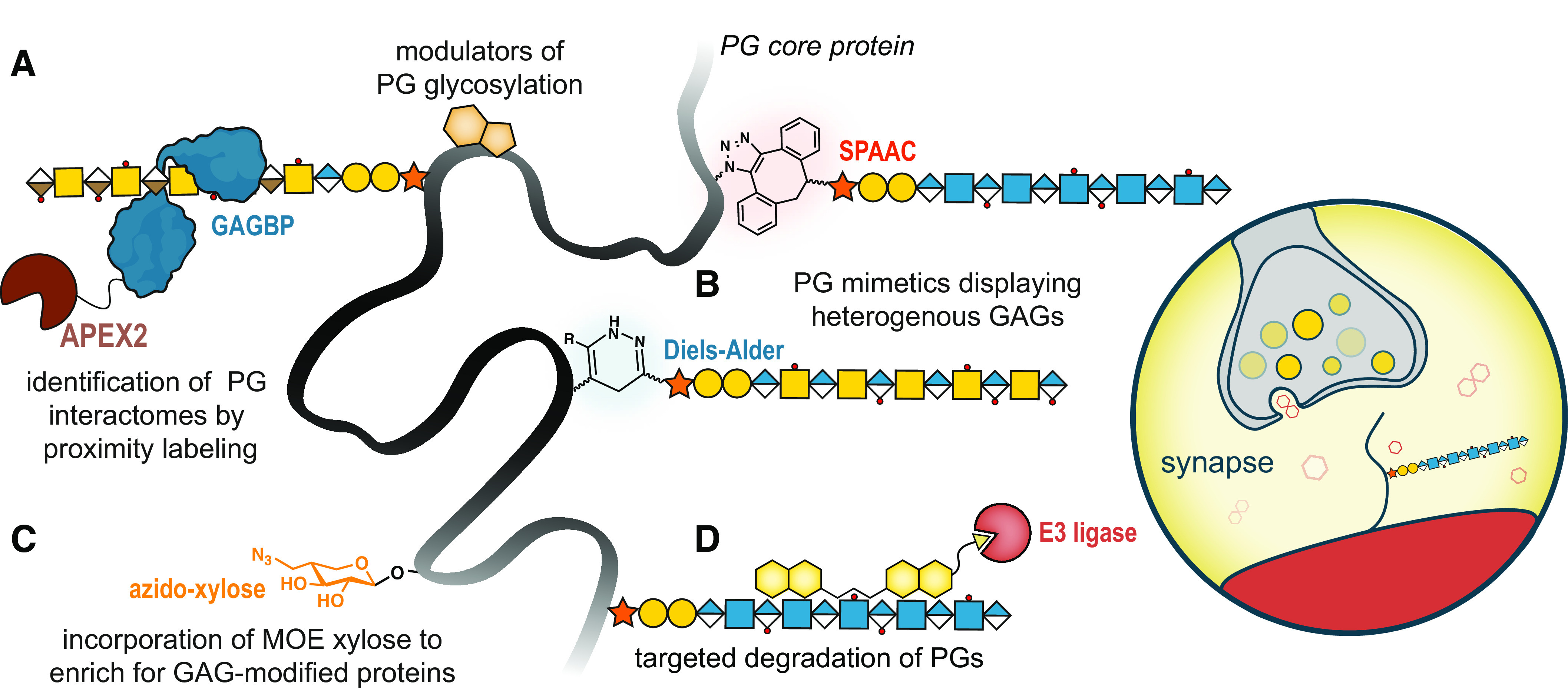
Uncharted opportunities in exploring proteoglycan structure-function relationships. A: the usage of proximity labeling approaches (e.g., APEX2, depicted) permits the identification of the collection of proteoglycans within a cell using GAGBPs. On the contrary, APEX2-PG fusion proteins can allow the identification of both GAG- and core protein-dependent interactions. Furthermore, these interactors can be derivatized to create small molecule modulators of PG glycosylation. B: the expansion of PG mimetics to include both HS and CS/DS GAGs will progress de novo PGs toward replicating native hybrid PGs, further enabling exploration of structure-activity relationships. C: xylose is a unique sugar in mammalian cells, primarily found in GAG core tetrasaccharide structures (GlcA-Gal-Gal-Xyl). The creation of a derivative with a functional handle for enrichment (e.g., azide, depicted) could allow for pull-down of GAG-modified proteins, including crucially, part-time PGs. D: through derivatization of PGs/GAGBPs or small molecules, molecular glue technologies could be used to modulate PG expression or structure in vitro. This could include the targeted degradation of disease-associated PG glycoforms using proteolysis targeting chimeras that recruit E3 ligase (depicted), or modulation of structure by recruiting GAG modifying enzymes (e.g., sulfatases to remove sulfate groups, or heparanase to cleave HS chains). The combination of these approaches would allow for a global view of the complete proteoglycan repertoire and characterization of their interactions and molecular mechanisms within a chosen biological setting, such as synapses. CS/DS, chondroitin sulfate/dermatan sulfate; GAGs, glycosaminoglycans; GAGBPs, GAG-binding proteins; HS, heparan sulfate; MOE, metabolic oligosaccharide engineering; PGs, proteoglycans.
