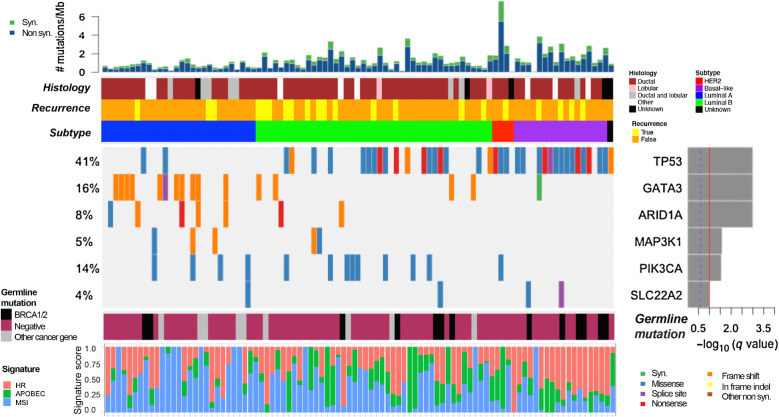
Figure 1.
Significant SNVs, short indels, and signature analysis. Comutation plot showing recurrent somatic alterations in significantly mutated genes across the cohort (N = 93) as analyzed by MutSig2CV. TP53, GATA3, ARID1A, MAP3K1, PIK3CA, and SLC22A2 are significantly mutated. The P values were computed using the Fisher method and truncated product method. FDR (q values) were generated using the Benjamini–Hochberg method to correct for multiple hypotheses. Genes that have a −log10 q-value ≥1 (red line) are considered significant. Bar graph (top) depicts the TMB (mutations/megabase) of each patient's tumor samples, followed by clinical annotations depicting histology, disease recurrence, and breast cancer subtype (key to the right of panel). Bottom panel annotations show cancer-specific pathogenic germline variants, and somatic mutational signatures of homologous recombination (HR) deficiency, APOBEC activity, and microsatellite instability (MSI).