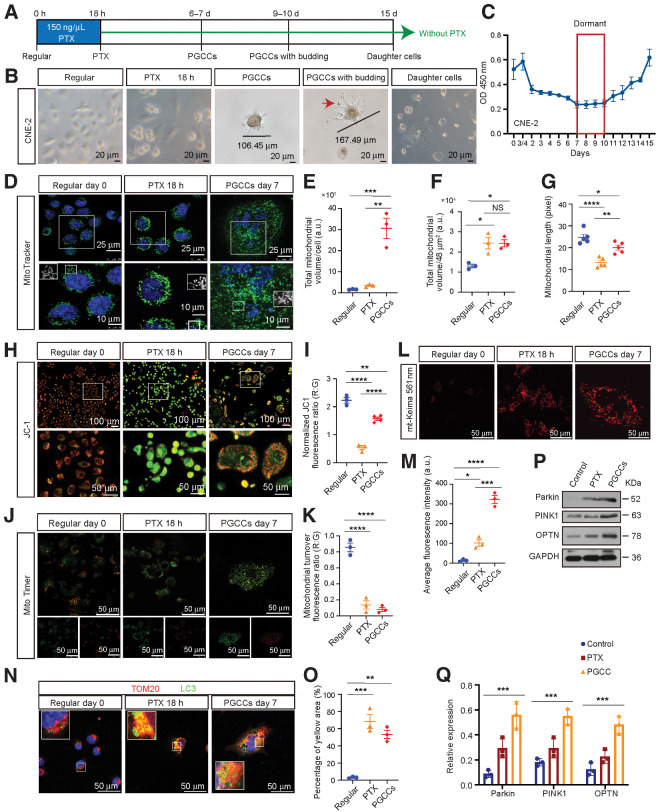
Figure 1.
Morphologic characteristics, mitochondrial morphology, and function analysis of PGCCs. A, Experimental design for PGCC induction and daughter cell generation in vitro. B, Light phase contrast microscopy images of cells at the indicated time points. Arrow, the mononuclear daughter cells. C, The CCK8 assays were performed to measure cell proliferation. O.D., optical density. D, Mitochondrial morphology was visualized using MitoTracker. E and F, Mitochondrial volume/cell was measured in three fields per group, with three independent replicates. a.u., arbitrary units. G, Mitochondrial length was measured in five fields per group, with three independent replicates. H, Confocal microscopy of mitochondrial membrane potential change with JC-1. Red puncta, maintained mitochondria; green puncta, depolarized mitochondria. I, The quantification of JC-1 results. J, Mitochondrial turnover visualized by MitoTimer transfection and labeling. K, The histogram shows the ratiometric quantification of three independent experiments of 30 cells per group. R:G, red:green. L, Fluorescent images of cells transfected with plasmid mt-Keima (mitophagy reporter). M, Quantification of average fluorescence intensity, with three independent replicates of 30 cells per group. N, IF costaining of TOM20 and LC3. Blue, nucleus; red, TOM20; green, LC3. O, Quantification of the percentage of yellow area. P, Western blot analysis. Q, Gray value analysis of Western blot. All data represent the mean ± SEM of at least three independent experiments. P values were calculated using one-way ANOVA. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; NS, not significant. PTX, paclitaxel.