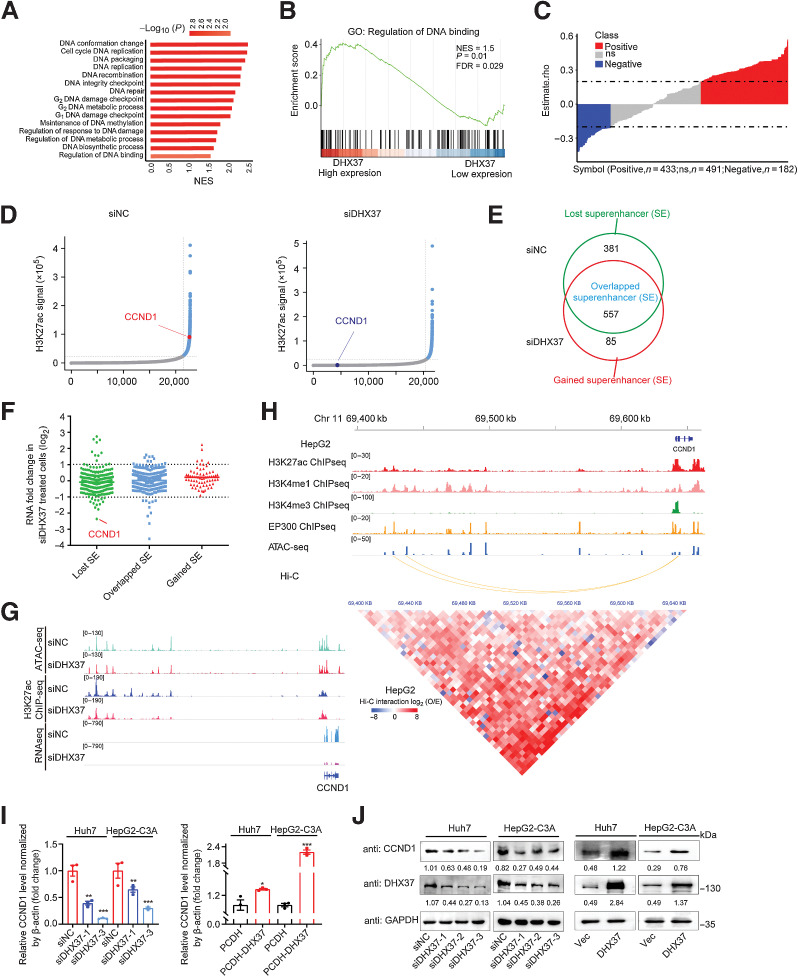
Figure 2.
DHX37 is strongly associated with transcriptional regulation and enhancer activity in HCC. A, GSEA of the biological processes associated with DHX37 expression in TCGA-LIHC cohort. The vertical axis indicates the GSEA biological process, and the horizontal axis indicates the NES of the biological process. The colors on the bar indicate the significance of enrichment. B, GSEA visualization of the regulation of DNA binding in TCGA-LIHC cohort. C, Correlation analysis of the expression of DHX37 and eRNAs. Each column represents an eRNA. The height of the column indicates the correlation between the expression of each eRNA and that of DHX37, and the color of the column indicates the significance of the correlation. Red, positive correlation (n = 433); blue, negative correlation (n = 182); gray, no significant correlation (n = 491). D, Hockey stick plots on the basis of input-normalized H3K27ac signals in Huh7 cells after silencing of DHX37. The enhancers are rank ordered as a percentage of the total signal. Superenhancers are obtained on the basis of the difference in the strength of the binding levels of molecular markers of enhancer transcriptional activity. Superenhancers are highlighted in blue with the ranks of selected superenhancer-associated genes. E, Definition of lost superenhancers (only in the siNC group), overlapped superenhancers (in both the siNC and siDHX37 groups), and gained superenhancers (only in the siDHX37 group). F, Gene expression changes as measured by RNA-seq of lost superenhancers, overlapped superenhancers, and gained superenhancers in siNC and siDHX37 Huh7 cells. The individual points are represented as mRNA fold change for superenhancer-associated genes. G, ATAC-seq, ChIP-seq, and RNA-seq profiles of the CCND1 locus in Huh7 cells treated with siNC and siDHX37. H, Epigenetic features of the CCND1 locus in HepG2 cells. A 5 kb resolution heatmap for chromosome 11 (hg38: 69400000–69645000 bp) in HepG2 cells showed the location of topologically associating domains, which was derived from ENCODE and reanalyzed on the basis of Juicer and hiccups software (bottom). The Hi-C interaction between promoters and superenhancers of CCND1 was evaluated by the O/E value, which was calculated as the observed value (estimated with normalized mapped reads) divided by the expected value. I and J, Relative mRNA and protein expression levels of CCND1 upon knockdown and overexpression of DHX37 in Huh7 and HepG2-C3A cells (n = 3). The values are expressed as the mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by one-way ANOVA or two-sided Student t test.