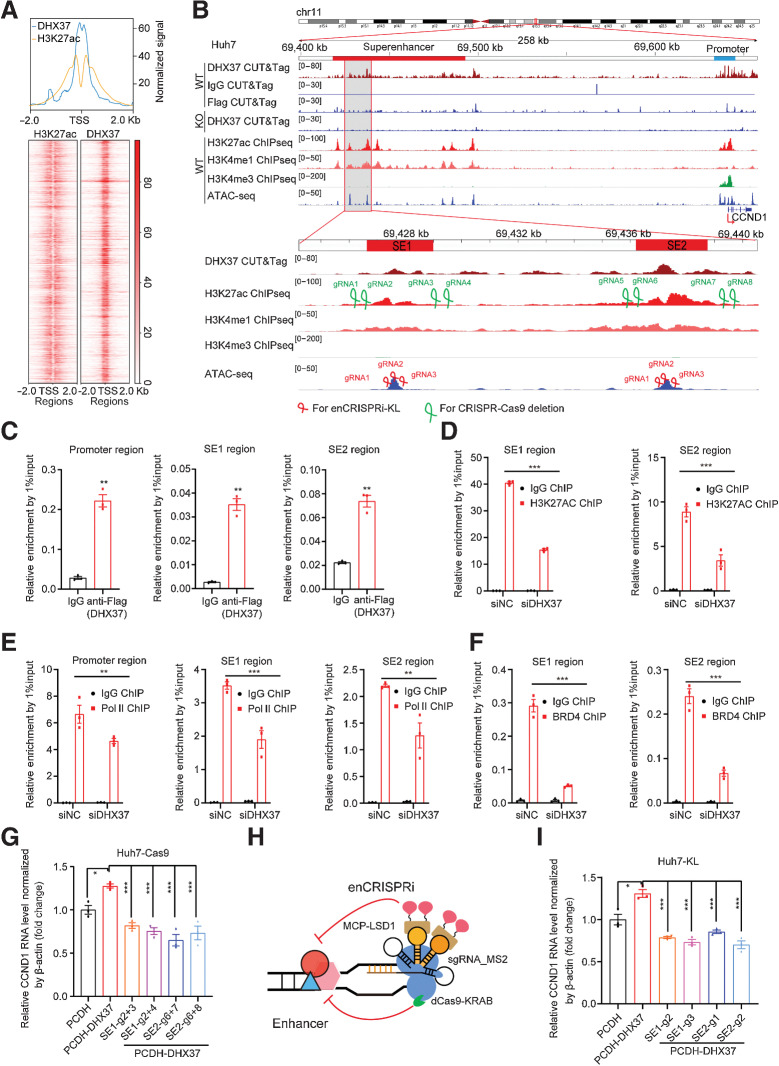
Figure 3.
CCND1 is a direct target gene regulated by DHX37 in a superenhancer-driving manner in liver cancer cells. A, Top, line plots showing CUT&Tag and ChIP-seq signals of DHX37 and H3K27ac centered at the summit of DHX37 peaks in Huh7 cells. Bottom, heatmap of CUT&Tag and ChIP-seq signals for DHX37 and H3K27ac [±2,000 bp windows around the center of the transcriptional start site (TSS)]. B, Profiles of DHX37, H3K27ac, H3K4me1, and H3K4me3 occupancy and ATAC-seq peaks at the CCND1 promoter (blue line) and superenhancer (red line) regions in wild-type (WT) and DHX37 knockout (KO) Huh7 cells. Gray shading indicates the occupancy of DHX37, which contains two specific constituent superenhancers (SE1 and SE2). sgRNAs were designed on the basis of the above peak. C, ChIP-qPCR analysis of DHX37 enrichment at the promoter and superenhancer regions of CCND1 (n = 3). D, Treatment with siDHX37 significantly reduced H3K27ac enrichment at the CCND1 superenhancer region compared with that in the siNC group (n = 3). E, Treatment with siDHX37 significantly reduced RNA Pol II occupancy at the CCND1 promoter and superenhancer regions compared with that in the siNC group (n = 3). F, Treatment with siDHX37 significantly reduced BRD4 enrichment at the CCND1 superenhancer region compared with that in the siNC group (n = 3). G, Relative mRNA levels of CCND1 upon overexpression of DHX37 and knockout of the SE1 and SE2 regions by CRISPR-Cas9 gene editing (n = 3). H, Schematic of the enCRISPRi system containing the dCas9-KRAB fusion protein, an sgRNA with two MS2 hairpins, and the MCP-LSD1 fusion protein. I, Blockade of CCND1 superenhancer regions by four individual sgRNAs significantly reversed the upregulated expression of CCND1 caused by DHX37 (n = 3). The values are expressed as the mean ± SEM (C–F and G–I). *, P < 0.05; **, P < 0.01; ***, P < 0.001 by one-way ANOVA and two-sided Student t test.