Abstract
Developing affordable and easily manufactured SARS-CoV-2 vaccines will be essential to achieve worldwide vaccine coverage and long-term control of the COVID-19 pandemic. Here the development is reported of a vaccine based on the SARS-CoV-2 receptor-binding domain (RBD), produced in the yeast Pichia pastoris. The RBD was modified by adding flexible N- and C-terminal amino acid extensions that modulate protein/protein interactions and facilitate protein purification. A fed-batch methanol fermentation with a yeast extract-based culture medium in a 50 L fermenter and an immobilized metal ion affinity chromatography-based downstream purification process yielded 30–40 mg/L of RBD. Correct folding of the purified protein was demonstrated by mass spectrometry, circular dichroism, and determinations of binding affinity to the angiotensin-converting enzyme 2 (ACE2) receptor. The RBD antigen also exhibited high reactivity with sera from convalescent individuals and Pfizer-BioNTech or Sputnik V vaccinees. Immunization of mice and non-human primates with 50 µg of the recombinant RBD adjuvanted with alum induced high levels of binding antibodies as assessed by ELISA with RBD produced in HEK293T cells, and which inhibited RBD binding to ACE2 and neutralized infection of VeroE6 cells by SARS-CoV-2. Additionally, the RBD protein stimulated IFNγ, IL-2, IL-6, IL-4 and TNFα secretion in splenocytes and lung CD3+-enriched cells of immunized mice. The data suggest that the RBD recombinant protein produced in yeast P. pastoris is suitable as a vaccine candidate against COVID-19.
Abbreviations: RBD, Receptor-binding domain; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; NHP, non-human primates; CHO, Chinese hamster ovary cells, HEK293T, human embryonic kidney cells; IMAC, immobilized metal ion affinity chromatography; RP, reversed-phase chromatography; CD, circular dichroism; VNT, viral neutralization titer; ESI-MS, Electrospray ionization tandem mass spectrometry
Keywords: SARS-CoV-2, P. pastoris, COVID-19, RBD, Subunit vaccine
Graphical Abstract
1. Introduction
Mammalian cell expression systems such as human embryonic kidney cells (HEK293T) are preferred for the production of complex therapeutic proteins due to their ability to introduce post-translational modifications identical or similar to those found in humans [1]. However, within the resource-poor contexts of low-income countries, these platforms are beset by issues of technological complexity, high operating costs and challenges such as the possibility of viral contamination in large-scale cultures [2]. An alternative is the use of yeast, such as Pichia pastoris (Komagataella phaffii). P. pastoris exhibits many advantages over mammalian cells regarding the simplicity and cost of culture media, growth rate and ease of genetic manipulation, while sharing many of the characteristics of their protein folding and secretion processes [2]. Growth on methanol as sole carbon source induces in P. pastoris the strong and tightly regulated AOX1 promoter, which can be employed to drive heterologous gene expression [3] and, taking advantage of secretion signals such as those of the alpha mating factor, can be used to obtain secreted recombinant proteins directly in culture supernatants with low levels of host contaminant proteins [4].
The RBD of SARS-CoV-2 is a glycosylated 25 kDa protein domain spanning residues N331-K529 of the spike protein, including eight cysteine residues forming four disulfide bonds. RBD mediates cell entry through the ACE2 host receptor and the levels of RBD-binding antibodies strongly correlate with neutralizing antibodies titers in convalescents [5]. The domain contains two glycosylation sites (N331 and N343) and a central twisted anti-parallel β-sheet formed by five strands connected by short helices and loops [6]. Glycosylation plays a key role in the immunogenicity and stability of the RBD protein [7]. Possibly for this reason, the glycosylation introduced by P. pastoris being more distant from mammalian cells could contribute to the protein’s immunogenicity [8].
The SARS-CoV-2 RBD with reduced glycosylation has been produced by others at high levels in P. pastoris as a suitable vaccine candidate against COVID-19 [3], [9], [10], [11], [12], [13]. Comparison by CD and tryptophan fluorescence between RBD from P. pastoris and HEK293T mammalian cells showed that the proteins were properly folded as well as having similar temperature stabilities, despite differences in glycosylation of the two expression platforms [3].
Here, the design of an RBD protein vaccine candidate is reported, with its production in P. pastoris, purification, physico-chemical characterization, capacity to elicit ACE2 receptor binding inhibitory antibodies and neutralizing responses in rodents and monkeys. The approach differs from the previously reported production of RBD in P. pastoris [3] by the inclusion of N- and C-terminal extensions aimed at modulating potential protein-protein interactions, and by optimizing the protein fermentation and purification processes.
2. Materials and methods
2.1. Biological reagents, protein designations and serum panels
Human ACE2 receptor chimeric proteins fused to a human or murine Fc antibody domain (hFc-ACE2, mFc-ACE2) as well as RBD fused to human Fc (hFc-RBD) were supplied by the Center of Molecular Immunology (CIM, Havana, Cuba). The chimeric proteins were purified by protein-A affinity chromatography (GE Healthcare Bio-Sciences, Uppsala, Sweden) from supernatants of stably transduced HEK293T cells and eluted with glycine 100 mM pH 3 (Merck, Darmstadt, Germany). hFc-RBD was conjugated to horseradish peroxidase (HRP) (Sigma-Aldrich, St. Louis, MO, USA) and designated hFc-RBD-HRP. H6-RBD refers to an N331-S531 RBD carrying an N-terminal His(6) tag produced as inclusion bodies in E. coli [14], while RBD-H6 refers to an N331-K529 RBD carrying a C-terminal His(6) tag, secreted into the supernatant of stably transduced HEK293T cells. Both proteins were purified by immobilized metal ion affinity chromatography (IMAC) (GE Healthcare Bio-Sciences, Uppsala, Sweden) and the final buffer was exchanged to phosphate buffered saline (PBS). The engineered RBD constructs described in the present work carry an amino-terminal segment denominated C-tag, and a carboxy-terminal six histidine tag (H6), and are referred to as C-RBD-H6, with the suffix PP or HEK describing the host (P. pastoris or HEK293T cells) in which they were produced.
The panel of human sera used as controls included sera from volunteers vaccinated with Pfizer/BioNtech [15] or Gamaleya’s Sputnik V (Gam-COVID-Vac) vaccine [16], and sera from convalescent patients. All individuals gave written informed consent for use of their serum.
2.2. Construction of Pichia pastoris strains producing SARS-CoV-2 RBD (C-RBD-H6 PP)
A sequence encoding residues 331–529 of the spike protein of SARS-CoV-2 strain Wuhan-Hu-1 (NCBI Acc. No. YP_009724390) with the N- and C-terminal extensions was codon-optimized for Saccharomyces cerevisiae, using J-Cat [17] and cloned in-frame with the KEX2 cleavage site of the pre-pro MATα sequence of pPICZαA (Invitrogen, Waltham, MA, USA), placing it under transcriptional control of the P. pastoris AOX1 promoter. Codon usage was optimized for S. cerevisiae because expression of C-RBD-H6 was initially attempted in both hosts and the codon usage patterns of both are similar [18], [19]. After sequence verification by Sanger sequencing using primers flanking the C-RBD-H6 gene (Macrogen, Seoul, South Korea), the expression plasmid was linearized with Sac I (New England Biolabs, Ipswich, MA, USA) and used to transform P. pastoris strain X-33 [20]. Following incubation for about 96 h at 28 ºC on YPD-zeocin medium (1 % yeast extract (Condalab, Torrejón de Ardoz, Spain), 2 % peptone (Condalab), 2 % glucose (Merck, Darmstadt, Germany), 2 mg/mL zeocin (Merck) 100 recombinant colonies were randomly picked and used to prepare frozen glycerol stocks.
For expression analysis, aliquots from the frozen stocks were grown in 800 µL of buffered glycerol complex medium [BMGY, 2 % (w/v) peptone (Condalab), 1 % (w/v) yeast extract (Condalab), 1.34 % (w/v) Yeast Nitrogen Base (Difco, Detroit, MI, USA), 0.4 µg/mL biotin (DC Fine Chemicals, Barcelona, Spain), 1 % (v/v) glycerol (Tecsiquim, Toluca, Mexico), 261 mM K2HPO4 (DC Fine Chemicals) and 739 mM KH2PO4 (Merck)] in 24-well deep plates at 28 °C, 300 rpm for 24 h. Then, the cells were collected by centrifugation, resuspended in buffered methanol complex medium [BMMY, same as BMGY but substituting 0.5 % (v/v) methanol (Merck) for glycerol (Tecsiquim)] and further cultured under the same conditions for 96 h. The clone exhibiting the highest expression of C-RBD-H6 PP was selected by dot-blot immunodetection and densitometric analysis of culture supernatant, and used to prepare working cell banks.
2.3. Fermentation
Fermentation was carried out in a 75-liter Chemap fermenter (Chemap, Volketswil, Switzerland) with a working volume of 50 L of culture medium [21]. Four Petri dishes were seeded from a frozen vial of a working cell bank, and incubated for 60–84 h at 30 °C. 8–10 isolated colonies were used to inoculate 2 L Erlenmeyer flasks, each containing 500 mL of basal salts medium [5 g/L yeast extract (Condalab), 15 g/L (NH4)2SO4 (Merck), 36 g/L glycerol (Tecsiquim), 40 mg/L histidine (Merck), 7.75 g KH2PO4 (Merck) or 5 g/L K2HPO4 (DC Fine Chemicals), 0.2 g/L CaCl2 (Scharlab, Barcelona, Spain), 1 mL/L of a trace elements solution: 6 g/L CuSO4·5H2O (Merck), 0.415 g/L KI (Merck), 3 g/L MnSO4·H2O (Merck), 0.1 g/L H3BO3 (Merck), 1 g/L MoNa2O4·2H2O (Scharlab), 20 g/L ZnSO4·7H2O (Merck), 65 g/L FeSO4·7H2O (Scharlab)] and 10 mL/L H2SO4 (Merck) and 2.5 mL/L of vitamin solutions [1.6 g/L biotin (DC Fine Chemicals), 8 g/L myo-inositol (Merck), 0.8 g/L calcium pantothenate (DC Fine Chemicals), 0.8 g/L pyridoxal hydrochloride (DC Fine Chemicals), 0.8 g/L thiamine di-hydrochloride (Sigma-Aldrich), 0.2 g/L nicotinic acid (Merck), 4 g/L K2HPO4 (DC Fine Chemicals), and 6.2 g/L KH2PO4 (Merck)], which were then incubated for 20–28 h at 30 °C and 250 rpm. The fermentation run was started by pooling the shake flask cultures and inoculating the fermenter with 2.7 L of this inoculum, with a starting pH of 5.0, regulated by pumping liquid ammonia (Merck) and a temperature of 30 °C. The fed-batch phase started, once dissolved oxygen increased, by adding 50% glycerol (Tecsiquim) at 540 mL/h for 2–3 h. Temperature was lowered to 25 °C and pH was increased to 6.0 one hour after starting the fed-batch phase. At the end of this phase, 800 mL of methanol were added at 60 mL/min flow rate of a Watson Marlow 520 peristaltic pump (Watson Marlow, Wilmington, MA, USA), and once the cells were adapted to this new carbon source, methanol was added first at 6 mL/L/h, then at 9 mL/L/h 4 h later, and at 12 mL/L/h when cell density reached 200 g/L. This last flow was maintained until the end of fermentation (38–44 h).
2.4. Purification of C-RBD-H6 PP
After 48 h of fermentation, the culture was harvested, and cells were removed by continuous centrifugation with a retention time of 5–10 min at 21,420 RCF, 4 oC. The resulting supernatant was filtered sequentially through 8 µm, 3 µm and 0.45 µm cellulose filters (Merck), and then concentrated and buffer-exchanged against PBS containing 5 mM imidazole (Merck) by tangential flow filtration with a 30 kDa Hydrosart® membrane (Sartorius, Göttingen, Germany). The conditioned sample was loaded onto a Chelating Sepharose™ FF colum (Cytiva, Marlborough, MA, USA) with cross-linked 6 % agarose beads modified with iminodiacetic (IDA) matrix (Cytiva) charged with Cu2 + and equilibrated in the same buffer, washed sequentially with 30 column volumes of PBS containing 10 mM and 20 mM imidazole (Merck), and eluted with 250 mM imidazole in PBS. The eluted protein was further purified on a 50 × 250 mm RP C4 column (Tosohaas, Tokyo, Japan) with a resin volume of 500 mL and a particle size of 15–20 µm, coupled to a Shimadzu LC-20AP semi-preparative HPLC purification system (Shimadzu, Kyoto, Japan). The column was equilibrated with solution A [0.05% TFA (Merck) in water] and the protein was eluted with 40 min linear gradient of 32–45% of solution B [0.1% TFA in acetonitrile (Merck)]. The protein eluted at 35–37% of solution B, approximately in one column volume. The fractions containing the protein were diafiltered vs. 20 mM Tris-HCl pH 7.4 with a 10 kDa Hydrosart® membrane (Sartorius), pooled aseptically, filtered through a 0.22 µm membrane (Merck) and stored at − 20 oC.
2.5. Analysis of C-RBD-H6 PP by ESI-MS
100 µg of purified C-RBD-H6 PP were deglycosylated with 500 units of PNGase-F (New England Biolabs, Ipswich, MA, USA) for 2 h at 37 oC in the presence of 0.5 M guanidine hydrochloride and 5 mM N-ethylmaleimide. A 10 μg aliquot of the resulting preparation was desalted in a C18 ZipTip (Merck Millipore, Burlington, VT, USA) and loaded into a metal-coated nanocapillary for ESI-MS analysis. The remainder was digested following a previously reported in-solution buffer-free trypsin digestion protocol [22] adapted to the analysis of SARS-CoV-2 RBD proteins that provides full-sequence coverage of the tryptic peptides and detection of post-translational modifications in a single ESI-MS spectrum [14]. Other experimental conditions for ESI-MS analysis were similar to those reported previously [14].
2.6. Surface plasmon resonance (SPR)
The interaction between mFc-ACE2 and C-RBD-H6 PP was studied by SPR in a BIACORE X unit (GE Healthcare, Tokyo, Japan) at 25 °C in multi-cycle mode. Briefly, mFc-ACE2 was immobilized on flow cell 1 (FC1) of a Protein A biosensor chip (GE Healthcare, Amersham, UK) following manufacturer’s instructions, and flow cell 2 (FC2) was used as the reference cell for background binding affinity. The real-time response of C-RBD-H6 PP over immobilized mFc-ACE2 was recorded in duplicate across a concentration range of 15–2000 nM, at a flow rate of 10 µL/min for 120 s, while the dissociation took place for another 120 s. The running buffer was PBS (pH 7.2). After each cycle the chip was regenerated using glycine buffer pH 2.0. The equilibrium dissociation constant (KD) was estimated with BIAevaluation® software (GE Healthcare, Tokyo, Japan) using the Langmuir 1:1 interaction model. At least five curves were taken into account for the calculation of kinetic parameters.
2.7. Animals and immunization schedules
Three different animal species were used to evaluate the immunogenicity of C-RBD-H6 PP: BALB/c mice, Sprague–Dawley (SD) rats, and African green monkeys (Chlorocebus aethiops sabaeus). The experimental protocols were approved by the Ethical Committee on Animal Experimentation of the Center for Genetic Engineering and Biotechnology (CIGB, Havana, Cuba) and the Center for Production of Laboratory Animals (CENPALAB, Bejucal, Cuba).
The immunization of African green monkeys was of animals aged 3–6 years weighing 2–7 kg, kept at the animal experimentation facility of CENPALAB, Cuba. The immunogen contained, per 500 µL: 50 µg of C-RBD-H6 PP protein with 0.3 mg of aluminum hydroxide gel as adjuvant (Alhydrogel ®, Brenntag Biosector, Denmark), in phosphate buffer (0.28 mg Na2HPO4, 0.31 mg NaH2PO42H·2O, 4.25 mg NaCl). Twenty individuals were randomly assigned to 3 groups: placebo (2 animals/gender, total 4), low dose (50 µg/dose, 3 animals/gender, total 6), and high dose (100 µg/dose, 5 animals/gender, total 10). The animals were immunized intramuscularly on days 0, 14 and 28. Seven days after the first and second boost, the animals were fasted overnight, sedated with ketamine hydrochloride (10 mg/kg, i.m.) (Liorad, Havana, Cuba) and bled from the femoral vein.
Procedures for mouse and rat immunization are described in Supplementary Material S1.
2.8. Evaluation of serum antibodies
Antibody detection by Enzyme Linked Immunosorbent Assay (ELISA).
The reactivity of sera from immunized animals was determined by ELISA. Briefly, the wells of 96-well microtiter plates (Corning Costar, Acton, USA) were coated overnight at 4 °C with 0.25 µg of RBD-H6 (produced in HEK293T cells) in 0.1 M sodium carbonate buffer (pH 9.6). After blocking the plates with 2 % skim milk (Oxoid, Basingstoke, UK), 0.05 % Tween 20 (Merck, Darmstadt, Germany), serially diluted animal sera or control monoclonal antibodies SS-1, SS-4, SS-7 and SS-8 (CBSSRBD-S.1, CBSSRBD-S.4, CBSSRBD-S.7 and CBSSRBD-S.8; Center for Genetic Engineering and Biotechnology, Sancti Spiritus, Cuba) [23], were added and incubated at 37 °C for 2 h in 0.2 % skim milk, 0.05 % Tween 20 in PBS, followed by six washes with PBS containing 0.05 % Tween 20. Bound antibodies were detected with anti-human IgG conjugated to HRP (1:10,000; 109–035–098; Jackson ImmunoResearch, West Grove, PA, USA) at 37 °C for 1 h, followed by washing. The assay was developed with 3,3–5,5-tetramethylbenzidine and quantified at 450 nm in a microplate reader (BMG Labtech, Ortenberg, Germany). This ELISA was also used for the evaluation of RBD reactivity using sera from immunized animals, Pfizer-BioNTech or Sputnik V vaccinees, and COVID-19 convalescents.
Serum antibody titers were expressed in arbitrary units (AU) with reference to a SARS-CoV-2 neutralizing serum, a value of 1 corresponded to 5 times the optical density reading of the blank control. Monoclonal antibody SS-8 was used as the reference for mouse sera, and a hyperimmune polyclonal serum as the reference for rat serum samples. The polyclonal serum was obtained by pooling sera from 10 animals submitted to dose-repeated toxicology studies showing the highest virus neutralization titers (Geometric Mean above 1:2500). For nonhuman primates (NHP) antibody titers, serum from a convalescent subject with a high SARS-CoV-2 neutralization titer was used as the reference.
2.9. Plate-based RBD to ACE2 binding assay
A competitive ELISA was performed to determine the inhibitory activity of anti-RBD polyclonal sera on the binding of an hFc-RBD-HRP conjugate to hFc-ACE2-coated plates. Briefly, the wells of ELISA plates were coated with 0.25 µg of recombinant hFc-ACE2 as described above. Then, mixtures containing hFc-RBD-HRP conjugate and serial dilutions of the sera were pre-incubated for 1 h at 37 °C. 100 µL of the mixtures were added to each Fc-ACE2 coated well and further incubated for 90 min at 37 °C. Binding of the HRP-tagged RBD to the receptor was detected with 3,3–5,5-tetramethylbenzidine as substrate, reading the results at 450 nm. A similar assay was used to characterize the ability of C-RBD-H6 PP and C-RBD-H6 HEK to block the interaction of hFc-RBD-HRP with hFc-ACE2-coated plates.
2.10. Microneutralization of live SARS-CoV-2 virus in Vero E6
Neutralization antibody titers were determined by a traditional virus microneutralization assay (MN50) using SARS-CoV-2 (CUT2010–2025/Cuba/2020 strain). Virus neutralizing titers (VNT50) were calculated as the highest serum dilution at which 50% of the cells remained intact according to neutral red incorporation in the control wells (no virus added). For detailed procedure see Supplementary Material S2.
2.11. Cellular immune response
Long-term cellular immune responses were evaluated in BALB/c mice. Five animals per group received 25 µg of C-RBD-H6 PP or placebo in a 100 µL volume delivery subcutaneously at days 0, 14 and 35. Blood samples were drawn two weeks after the last immunization, and the animals were euthanized 3 months later to assess the response to an in vitro antigen recall in systemic (splenocytes) and lung-resident cells. The splenocytes were isolated by organ perfusion with gentamycin-supplemented RPMI1640 culture medium (Gibco, Invitrogen, Waltham, MA, USA), and lung cells were dissociated with the Miltenyi reagent set (130−095−927) in C-tubes (130−093−237), using an automated dissociator (Gentle MACS Octo dissociator, Miltenyi, Bergisch Gladbach, Germany). In both cases the remaining erythrocytes were lysed with ACK solution (A1049201, Gibco, Invitrogen, Waltham, MA, USA). Then, the splenocytes were resuspended in RPMI 1640, gentamycin 10 µg/mL, fetal bovine serum (FBS) 10% (Gibco) and directly used to study cellular response. Lung cells were suspended in a buffer for negative selection of CD3 + cells and further purified, after two washes, with the Pan T Cell Isolation Kit II (130–095–130, Miltenyi, Bergisch Gladbach, Germany), suspending the resulting CD3 + preparation in the same medium as the splenocytes. Live lung CD3 + enriched cells and splenocytes were counted with a flow cytometer (CyFlow, Sysmex, Norderstedt, Germany).
For the re-stimulation assays, splenocytes or lung CD3 + enriched suspensions were diluted to 107 CD3 + live cells/mL and 50 µL of each sample were seeded in two 96-well U bottom tissue culture plates. Cells were re-stimulated with 50 µL of 20 µg/mL C-RBD-H6 PP, or just medium (unstimulated) for 72 h, and the supernatant was analyzed at 1:2 dilution using specific Deluxe cytokine kits (BioLegend, San Diego, CA, USA) for IL-2 (43104), IFNγ (430815), IL-6 (431315), IL-4 (431106) and TNFα (430915) following manufacturer’s instructions. The cells were transferred at that point to anti-IFNγ-coated ELISpot plates (Mabtech, Stockholm, Sweden) and the results were analyzed 24 h later according to established procedures.
2.12. ELISpot assay with samples from previously infected individuals
Peripheral blood mononuclear cells (PBMCs) from COVID-19-convalescent subjects were isolated from 7 mL of whole blood collected into CPT tubes (Becton Dickinson, Franklin Lakes, NJ, USA), and stored in liquid nitrogen until analyzed. After resting cells overnight in Optmizer™ media (Gibco, Invitrogen, Waltham, MA, USA) CD3 + live cells were counted by flow cytometry and seeded on round-bottom plates (650160, Greiner Bio-One GmbH, Kremsmunster, Austria) at 5 × 104 cells per well with 10 µg/mL of C-RBD-H6 PP for 72 h. Cells were transferred to anti-IFNγ pre-coated plates (3420–4APW, Mabtech), and the numbers of IFNγ-secreting T cells were determined after 20 h of incubation. All individuals gave written informed consent for use of their samples.
2.13. Statistical analysis
Prism 8.4.3 software was used for statistical analysis. The normality of all datasets was assessed with the Shapiro-Wilk test. Normally distributed data were compared using the Student’s t-test for paired or unpaired samples, depending on experimental design. Non-normal data were compared with Mann-Whitney’s or Wilcoxon’s rank match-paired tests. Comparisons of more than two groups used Kruskal Wallis multiple analyses followed by Dunn’s post test. Spearman’s test was used to assess parameter correlations. Sigmoidal dose-response curves were transformed into a linear form using natural log transformation for dilutions and the function NORM.S.INV (Microsoft Excel) function for normalized OD450 nm data according to:
NORM.S.INV[(OD450nmSample-OD450nmMinimum)/(OD450nmMaximum-OD450nmMinimum)].
Paired comparison of slopes and X and Y intercepts, after adjusting data to a linear equation, indicated no significant differences between the results for each coating condition. Experimental designs included two replicates per sample and three independent experiments.
3. Results
3.1. Design of the C-RBD-H6 expression cassette
The protein denominated here denoted as C-RBD-H6 PP was designed as a potential subunit vaccine candidate against SARS-CoV-2. This protein has a modular structure consisting of a globular central -RBD- domain comprising residues N331-K529 of the spike protein, flanked by additional N- and C-terminal segments that contain polar and flexible linkers rich in Glycine and Serine (Gly9-Ser15 and Gly215-Ser229, sequence of C-RBD-H6 PP are shown in Supplementary Material S3). These extensions prevent potential protein-protein interactions and facilitate protein purification by ensuring that a C-terminal His(6) tag is well exposed. Both extensions are spatially well separated from the receptor binding motif, and their presence should limit, through steric hindrance, potential aggregation problems associated with the presence of exposed and disulfide-bonded Cys76 and Cys210.
3.2. Expression and purification of C-RBD-H6 PP
A construct for the expression in P. pastoris, under control of the AOX1 promoter, of protein C-RBD-H6 was prepared as described in Materials and Methods and designated pPICZα-CtagRBDH6. This construct was used to transform P. pastoris strain X-33 to obtain C-RBD-H6-producing clones. After screening for the most productive clone, a single strain denominated X33–23 was chosen for further work. This strain, when used in fermentation runs at a scale of 50 L, yielded a dry cell weight of 58.15 ( ± 14.54) g/L, and a C-RBD-H6 titer of 68.38 ( ± 15.70) mg/L (data averaged from 15 independent processes). SDS-PAGE and Western blotting profiles of culture supernatants from three separate C-RBD-H6 PP-producing clones, including X33–23, are shown in Supplementary Material S4.
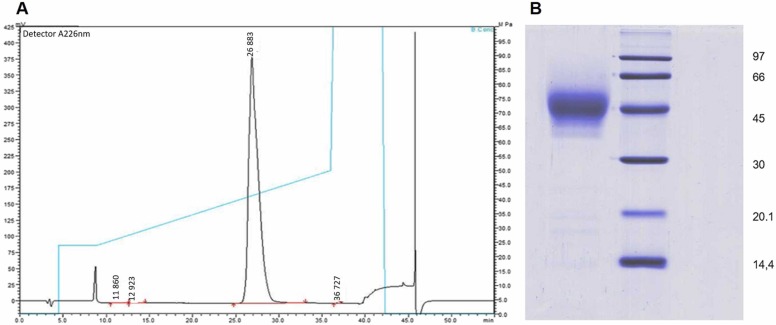
C-RBD-H6 PP protein was purified from fermentation supernatants of strain X33–23 by IMAC column charged with Cu2 + , and purified by RP. The entire process yielded 30–40 mg of pure C-RBD-H6 PP per L culture medium, with a purity equal to or higher than 98% ( Fig. 1).
Fig. 1.
RP-HPLC and protein electrophoresis. (A) Analysis of protein C-RBD-H6 PP on a reversed phase C8 Vydac analytical column. The gradient is shown by a blue line. Purity is at least 98.6%. (B) Coomassie Blue-stained 12.5% SDS-PAGE of 10 μg of purified C-RBD-H6 PP under reducing conditions. Lane 1: protein C-RBD-H6 PP; lane 2 molecular weight markers.
3.3. Structural analysis of C-RBD-H6 PP by ESI-MS
The sequence, N-glycosylation status and disulfide bonding pattern of C-RBD-H6 PP was examined by ESI-MS after deglycosylation with PNGase F and tryptic digestion. Full-sequence coverage of C-RBD-H6 PP was achieved, confirming the identity and integrity of the resulting protein (signal assignment from mass spectra is summarized in Table 1. ESI-MS/MS analysis of the signal detected at m/z 1399.64 (four charges) confirmed full N-glycosylation of Asn331 and Asn343 (the two N-glycosylation sites of the RBD within the context of the viral Spike protein), as they were transformed into Asp residues by the action of PNGase F (see underlined residues in Table 1). The four disulfide bonds (C336-C361, C379-C432, C391-C525 and C480-C488) present in the native S protein of SARS-CoV-2 were also detected in this ESI-MS spectrum (Table 1). Tryptic peptides containing free cysteine residues or S-S scrambling variants were not detected.
Table 1.
Summarized sequence verification of N-deglycosylated C-RBD-H6 PP from ESI-MS of tryptic peptides generated by in-solution buffer-free digestion.
| Codea) | m/zTheor | z | m/zExp | Assignment |
|---|---|---|---|---|
| C336-C361, Nt | 1399.64 | 4 | 1399.63 |
NWSFFSNIGGSSGGS-331DITNLCPFGEVFDATR346 b) / 358ISNCVADYSVLYNSASFSTFK378 (Native C336-C361, N-terminal end, Asn331 and Asn343→Asp) |
| C336-C361, Nt-NWSF | 1266.09 | 4 | 1266.07 |
FSNIGGSSGGS-331DITNLCPFGEVFDATR346 b) / 358ISNCVADYSVLYNSASFSTFK378 (Native C336-C361, N-terminal end -NWSF, Asn331 and Asn343→Asp corresponding to the two N-glycosylation sites) |
| F347-R355 | 557.28 | 2 | 557.26 | 347FASVYAWNR355 |
| K356-R357 | 303.21 | 1 | 303.20 | 356KR357 |
| C379-C432 | 1020.81 765.86 |
3 4 |
1020.79 765.85 |
379CYGVSPTK386 | 425LPDDFTGCVIAWNSNNLDSK444 (Native C379-C432) |
| C379 * -C432 | 982.71 | 4 | 982.69 |
64CYGVSPTK71 | 418IADYNYKLPDDFTGCVIAWNSNNLDSK444 (Native C379-C432, 1 missed cleavage) |
| C391-C525 | 992.52 | 4 | 992.47 |
387LNDLCFTNVYADSFVIR403 | 510VVVLSFELLHAPATVCGPK528 (Native C391-C525) |
| G404-R408 | 575.28 | 1 | 575.27 | 404GDEVR408 |
| Q409-K417 | 450.25 | 2 | 450.24 | 409QIAPGQTGK417 |
| I418-K424 | 443.72 | 2 | 443.71 | 418IADYNYK424 |
| V445-R454 | 609.80 | 2 | 609.78 | 445VGGNYNYLYR454 |
| L455-R457 | 435.27 | 1 | 435.26 | 455LFR457 |
| S459-R466 | 495.77 | 2 | 495.76 | 459SNLKPFER466 |
| C480-C488 | 1589.38 1192.29 |
3 4 |
1589.35 1192.26 |
467DISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTNGVGYQPYR509 |__________| (Native C480-C488) |
| Ct* -His6 | 799.68 600.01 |
3 4 |
799.67 600.00 |
529K-GGSGGSSSSSSSSSSIEHHHHHHb) (C-terminal end, 1 missed cleavage) |
| Ct-His6 | 756.98 567.99 |
3 4 |
756.97 567.98 |
GGSGGSSSSSSSSSSIEHHHHHHb) (C-terminal end) |
Nt: N-terminal end, Ct-His6: His-tag C-terminal end. C#-C# corresponds to tryptic peptides linked either by intermolecular disulfide bonds or a tryptic peptide that contains an intramolecular disulfide bond in its structures. m/zcalc correspond to the calculated m/z values for all tryptic peptides generated by the in-solution buffer-free digestion of the N-deglycosylated protein. m/zexp correspond to the experimental m/z values for all tryptic peptides observed in the ESI-MS analysis shown in Fig. 4.C. Regions of the sequence written in italics do not correspond to the RBD of SRAS-CoV-2 and were inserted in the cloning stage, while underlined residues indicate the conversion of N-glycosylated asparagines (Asn331 and Asn343) into aspartic acid residues by the action of PNGase-F (Asn331 and Asn343→Asp).
ESI-MS analysis of the deglycosylated C-RBD-H6 PP protein and its derived tryptic peptides confirmed that N-glycans in its structure increased considerably its molecular mass by SDS-PAGE analysis. Sugar content calculated in 12 protein batches was 42.7 % (40.7–43.8) relative to the molecular mass. The analysis of ESI-MS spectra is included as a Supplemental Material S5.
3.4. Characterization of binding affinity of C-RBD-H6 PP to ACE2 by Surface Plasmon Resonance
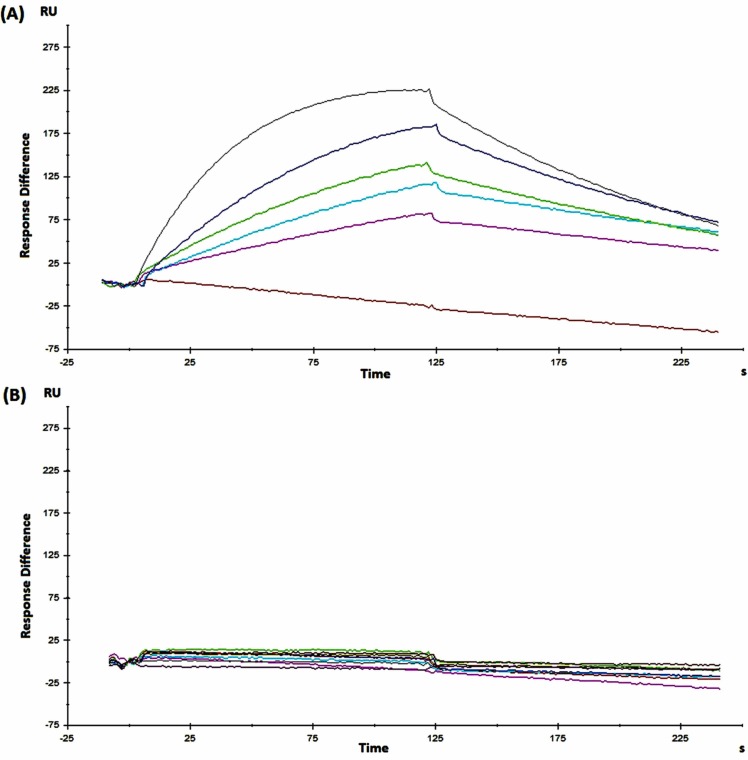
In order to study the affinity of the C-RBD-H6 PP/ACE2 interaction, mFc-ACE2 was immobilized via its Fc region onto a Biacore Protein A chip. This chip produced no appreciable signal in terms of response units (RU) with a non-related protein (negative control), as shown in Fig. 2. In contrast, C-RBD-H6 PP exhibited an association rate to mFc-ACE2 of 5.4 × 105 M-1·s-1 and a dissociation rate of 7.7 × 10-3 s-1. Equilibrium was reached after 25–30 s, with an estimated dissociation constant of KD = 14.3 × 10-9 M. The association/dissociation rates as well as KD were in the range previously reported in the literature for the RBD-ACE2 molecular interaction [24].
Fig. 2.
SPR analysis of the interaction of C-RBD-H6 PP with mFc-ACE2 in a single-cycle BIACORE experiment. (A) Sensorgrams corresponding to one of the replicates of protein C-RBD-H6 in PBS, pH = 7.2. (B) Non-related human recombinant Epidermal Growth Factor protein (also produced in yeast at CIGB, Havana, Cuba), used as negative control for the interaction with immobilized mFc-ACE2.
3.5. Secondary structure analysis by Circular Dichroism (CD) Spectroscopy
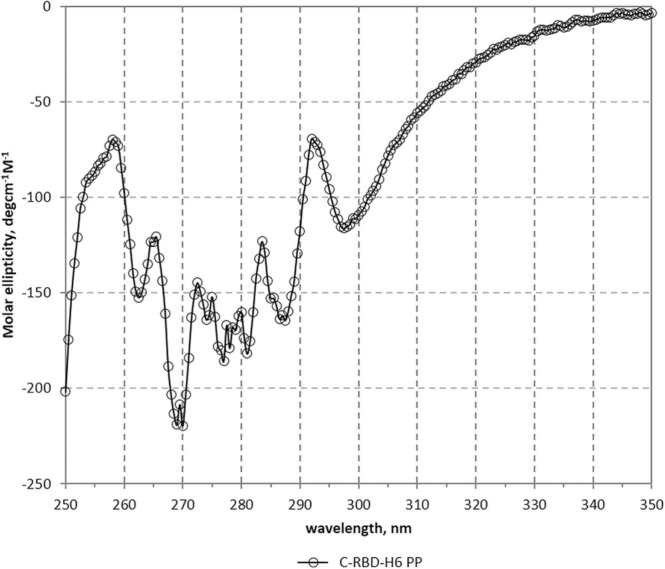
The far UV CD spectrum of C-RBD-H6 PP revealed characteristic bands similar to those previously reported for other recombinant RBD proteins [3], with maxima at 192 and 231 nm – due to the aromatic contribution - and a minimum at 207 nm (the procedures and CD spectrum are described in Supplementary Material S6). Furthermore, as shown in Table 2 the secondary structure content of the protein estimated by BeStSel (7.9% helix, 28.7% beta - antiparallel, relaxed and right-handed-, 12.9% turn and 50.5% others) was very similar to the values assigned using the 3D coordinates [6]. Moreover, as observed in Fig. 3, the near UV CD spectrum of the protein is well structured, with bands at 263, 269, 277, 281 and 299 nm, indicating the presence of well-packed aromatic and cysteine residues as expected of a correctly folded protein.
Table 2.
Secondary structure content of the C-RBD-H6 PP protein estimated by CD (BeStSel) and 3D coordinates (DSSP).
| Secondary Structure Element | Secondary Structure Content, % |
|
|---|---|---|
| Method | ||
| BeStSel | DSSP | |
| Helix | 7.9 | 9.3 |
| Beta antiparallel | 28.7 | 22.4 |
| Beta parallel | 0.0 | 0.0 |
| Turn | 12.9 | 22.4 |
| others | 50.5 | 45.9 |
| Helix1 (regular) | 3.3 | – |
| Helix2 (distorted) | 4.6 | – |
| Beta Antiparallel_1 (left-handed) | 0.9 | – |
| Beta Antiparallel_2 (relaxed) | 12.0 | – |
| Beta Antiparallel_3 (right-handed) | 15.8 | – |
Fig. 3.
Near UV CD spectrum of the C-RBD-H6 PP protein. Bands at 263, 269, 277, 281 and 299 nm, indicate the presence of well-packed aromatic and cystine residues.
3.6. Analysis of the antigenicity of C-RBD-H6 PP
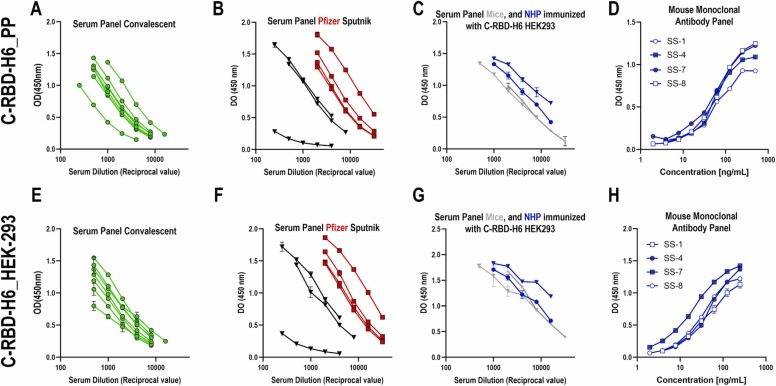
To confirm antigenicity of C-RBD-H6 recognition by known sera and monoclonal antibodies, the binding of the murine anti-RBD monoclonal antibodies SS-1, SS-4, SS-7 and SS-8 to C-RBD-H6 PP and C-RBD-H6 HEK was first compared by ELISA. These mAbs were obtained by immunization with RBD-H6 (produced in HEK293T cells), and one, SS-8, competes with ACE2 for binding to the RBD with an IC50 of 122.7 pM [23]. As shown in Fig. 4D,H, the reactivity of C-RBD-H6 PP to these four mAbs was indistinguishable from that of C-RBD-H6 HEK, except for SS-7 that shows increase recognition of the HEK derived protein.
Fig. 4.
Antigenicity of the RBD protein produced in P. pastoris. C-RBD-H6 PP (upper panel) or RBD-H6 HEK (lower panel) were used to coat ELISA plates. All immune sera and monoclonal antibodies were used in serial two-fold dilutions. (A,E) Sera from COVID-19 convalescents; (B,F) Sera from Pfizer/BioNTech (red squares) or Sputnik (black triangles) vaccinees; (C,G) Sera from mice (grey) or NHP (blue) immunized with CRBD-H6 HEK adjuvanted in alum-phosphate; (D,H). SS-1, SS4, SS-7 and SS-8 monoclonal antibodies obtained by immunizing mice with RBD-H6 HEK. Mean OD450nm ± SD is depicted for two replicates per experimental point. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Next, eight convalescent human sera with high neutralization titers against live SARS-CoV-2 as determined in Vero E6 cells, as well as eight sera from Pfizer-BioNTech or Sputnik V vaccinees were incorporated into the testing panels. A non-folded and non-glycosylated C-RBD-H6 protein produced as inclusion bodies in E coli BL-21 was used as a negative control. C-RBD-H6 PP protein displayed binding comparable to the protein purified from mammalian cells (Fig. 4A,E and B,F). Further characterization was conducted using polyclonal sera with known anti-SARS-CoV-2 neutralizing activity, obtained by immunization of mice and NHP with C-RBD-H6 HEK. Again, there were no significant differences between the reactivity of C-RBD-H6 produced in either system (yeast or mammalian) toward these polyclonal sera (Fig. 4 C,G).
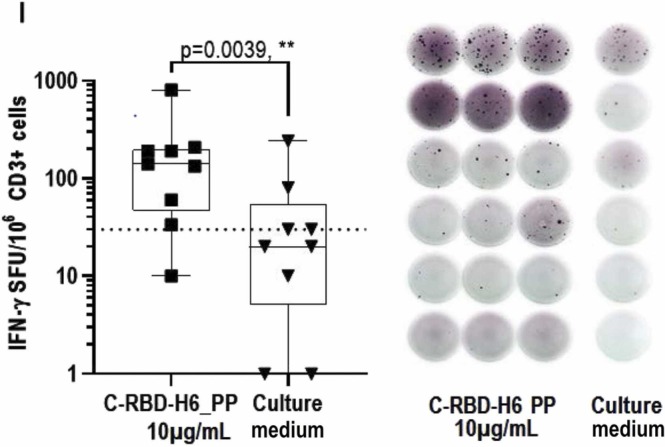
The C-RBD-H6 PP protein was also used to recall an in vitro a cellular response, measured by detecting IFNγ secretion using an ELISpot with PBMCs from COVID-19 convalescents sampled at least three months after hospital discharge. As shown in Fig. 5, stimulation with 10 µg/mL of the P. pastoris-produced protein induced IFNγ secretion from the PBMCs of COVID-19 convalescent subjects.
Fig. 5.
ELISpot with samples from COVID-19 convalescents. C-RBD-H6 PP stimulated INFγ secretion in CD3 + cells from naturally infected individuals. Comparison made by Wilcoxon’s matched paired test.
3.7. Functionality of C-RBD-H6 PP
3.7.1. C-RBD-H6 produced in P. pastoris elicited RBD-ACE2 receptor binding inhibitory and SARS-CoV-2 neutralizing antibodies in rodents and NHP
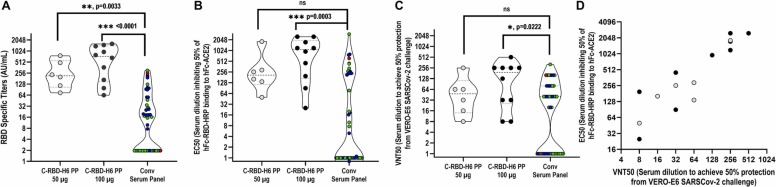
The immunogenicity of C-RBD-H6 PP was evaluated in NHP (green monkeys) using a short 0–14–28 days intramuscular administration schedule with two dose levels (50 µg and 100 µg). The results indicate the presence of a trend to a dose-response effect since seroconversion rates after the first booster were 83 % (5 out of 6 animals) and 100 % (all 10 animals) for the 50 and 100 µg dose levels, respectively, and 100 % after the second booster for both dose levels. Total IgG median titers increased to 216 (25–75 %: 105–558) AU/mL and 727 (25–75 %: 149–1410) AU/mL for the low and high-dose groups respectively. In both cases, the titers were significantly higher than those detected in the convalescent serum panel (p < 0.05, Kruskal-Wallis) ( Fig. 6A). Median ACE2 binding inhibition titers were 1:209 (25–75 %: 115–673) and 1:1081 (25–75 %: 169–1938) for the 50 µg and 100 µg dose groups, respectively, correlating well with the higher binding titers exhibited by the latter group. In this case, only the 100 µg dose group exhibited inhibitory titers higher than those of the convalescent panel (p < 0.001, Kruskal-Wallis) ( Fig. 7B). Similarly, the 100 µg dose group exhibited a median viral neutralization titer (VNT50) of 1:192 (25 %−75 %=26–156), which dropped to 1:48 (25 %−75 %=14–112) for the 50 µg dose group but was still at the level of the 1:40 (25 %−75 %= 1–80) VNT50 of the convalescent panel. Altogether, these data are consistent with a trend to a dose-related effect (Fig. 6). Specific IgG antibodies and RBD-ACE2 binding inhibition were detected only in C-RBD-H6 PP protein-inoculated animals and not in control animals. These antibody responses evidenced the existence of dose dependence and boosting effects, compared to the negative results obtained in control animals.
Fig. 6.
Immunogenicity of C-RBD-H6 PP in NHP. (A) Evaluation of RBD specific IgG in NHP immunized with doses of 50 µg (6 animals) or 100 µg (10 animals) of C-RBD-H6 PP, 14 days after the end of a 0–14–28 days intramuscular schedule. (B) Evaluation in NHP of EC50 for ACE2 binding inhibition. (C) Evaluation in NHP of EC50 for the PRNT in the microneutralization assay. (D) Association/correlation analysis of the ACE2 binding inhibition and microneutralization tests in NHP (Spearman, r = 0.8994, p < 0.0001).
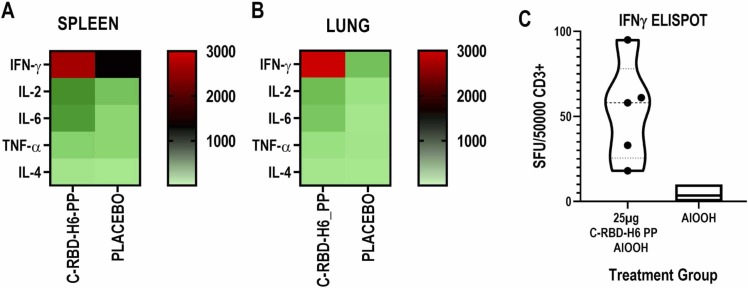
Fig. 7.
Heatmap of the cytokine response. (A) Splenocytes and (B) lung CD3 + -enriched cells, after restimulation with C-RBD-H6 PP. The cells were pooled from 4 to 5 mice per group, 3 months after the last immunization with 3 subcutaneous doses of 25 µg of C-RBD-H6 PP or placebo. Non-stimulated controls were subtracted from re-stimulated samples. (C) ELISpot assay of splenocytes stimulated with C-RBD-H6 PP.
Details of the immunization procedures of mice and rats and data on the immunogenicity, ACE2 binding inhibition and neutralizing titers of the resulting sera are available in Supplemental Material S1.
3.7.2. Cellular response
Cellular recall responses were evaluated three months after the last immunization of BALB/c mice receiving subcutaneous doses of 25 µg of C-RBD-H6 in alum at days 0–14–35. The presence of a memory response in the splenocytes of immunized animals was evidenced by a significant induction of IFNγ-secreting clones upon incubation with the C-RBD-H6 PP antigen (Fig. 7C). An analysis of the supernatant of recall reactions showed that the most strongly induced cytokine was IFNγ followed by IL-2, IL-6 and, to a lesser extent, TNFα and IL-4. This pattern was very similar between cells from the systemic compartment (Fig. 7A) and lung CD3+-enriched cells (Fig. 7B), demonstrating that cellular responses can also be recalled in the organ primarily affected during SARS-CoV-2 infections.
4. Discussion
Results from this study demonstrate a vaccine candidate based on C-RBD-H6 PP, an antigen derived from the RBD of the Spike protein of SARS-CoV-2, can produced in the yeast P. pastoris. The specific RBD sequence used in this candidate was selected based on state of the art evidence regarding the contribution of this domain to the induction of neutralizing antibodies [8], [25], [26], [27].
Optimal conditions for production of a recombinant protein in the P. pastoris expression system differ according to the target protein. For C-RBD-H6 PP we were able to obtain 30–40 mg/L RBD in a 50 L fermentation process with purity > 98%, which is close to the yield reported by others for a Pichia-produced RBD in a 7 L fermentation setup, although only 90% purity was reported [3]; high purity is an essential condition for a vaccine candidate. The Gly/Ser rich segments introduced in the genetic construction have been widely used in protein engineering as linkers for fusion proteins [28], This design was aimed in part to sterically preclude potential protein aggregation involving the protein surface sequence spatially close to the disulfide bridge Cys76-Cys210; Gly offers flexibility because it can adopt dihedral angles not possible for other amino acids; and Ser confers solubility as a hydrophilic amino acid.
Unlike other reports expressing the RBD in P. pastoris [29], [30], [31] here the potential N-glycosylation site at N331 in C-RBD-H6 PP was included. Although inclusion of N331 leads to more heterogeneous N-glycosylation [3], [32], [33], it should be noted that P. pastoris N-glycans are often hypermannosylated [34], [35], and mannosylation enhances the activation of antigen-presenting cells like macrophages and dendritic cells, increasing immunogenicity over that of non-glycosylated counterparts [8], [36], [37], [38]. In fact, others [29], [31] used a lipid-modified alum adjuvant or a saponin-based adjuvant, respectively, to overcome the poor immunogenicity of their N331-less RBD antigens. Another potential problem stemming from the absence of N331 is protein aggregation, detected by [29]. Again, the ionic interactions provided by the additional sugars attached to N331 may help to counteract this problem, which was absent in the case of C-RBD-H6 PP.
Physico-chemical analysis of C-RBD-H6 by mass spectroscopy demonstrated the presence of all four correctly formed disulfide bonds without scrambled species, and circular dichroism spectroscopy showed that its secondary structure makeup was compatible with that expected from the crystallographic structure of the spike trimer from SARS-CoV-2. The correct folding of C-RBD-H6 was further confirmed by functional assays such as its ability to bind to ACE2, as well as SPR assays in which it bound ACE2 with an affinity similar to that reported by others for this interaction [39], [40], [41]. Correct folding is one of the virtues of P. pastoris as an expression host, which combines the quality control mechanisms of the eukaryotic secretory pathway with the advantages of a microbial system [3], [42], and is a necessary requirement for the induction of an antibody response able to neutralize SARS-CoV-2 by blocking viral entry via inhibition of the RBD-ACE2 interaction.
Remarkably, despite the known differences between yeast and mammalian N-glycosylation, the reactivity of C-RBD-H6 PP was essentially identical to that of C-RBD-H6 HEK towards sera from mice and monkeys immunized with the latter, sera from COVID-19 convalescents, and sera from Pfizer-BioNTech or Sputnik V vaccinees. C-RBD-H6 PP was also able to stimulate cellular responses mediated by IFNγ secretion in lymphocytes isolated from individuals previously infected with SARS-CoV-2.
Immunization of mice, rats and NHP with C-RBD-H6 PP elicited antibodies that inhibited RBD-ACE2 receptor binding and were able to neutralize live SARS-CoV-2 in microneutralization tests. Even though the time constraints imposed by the accelerated development of this vaccine candidate required that the neutralizing activity of NHP sera be evaluated just one week after administration of the third dose, the resulting neutralizing titers were still higher than those of the convalescent serum panel used as a control. A longer period before evaluation might yield further improved results, allowing for the maturation and selection of B cell clones producing antibodies of higher avidity and, consequently, higher neutralizing titers.
Funding
This work was supported with funds from the BioCubaFarma, the Center for Genetic Engineering and Biotechnology, and by the Grant of the National Science and Technology Program - Biotechnology, Pharmaceutical Industry and Medical Technologies, of the Ministry of Science and Technology, project code PN385LH007-048. The Civilian Defense Scientific Research Center supported the microneutralization assays.
Author contributions
GGN: provided original ideas and study concept and design, data curation, analysis and interpretation of data, and drafting, review and editing of the final version of the manuscript and studies supervision. MLF: contributed to analysis and interpretation of data, study design, review of the manuscript and studies supervision. LJGL: contributed to ESI-MS study design, acquisition of data, analysis and interpretation of data, and drafting and review of the manuscript. LAER, IAM and YRG: performed the ESI-MS studies, acquisition of data, analysis and interpretation of data. GCH: performed glycosylation studies. ACR: performed drafting and execution of Biacore studies. GMP: performed the protein purification studies. MPI and JZS: performed fermentation studies. GCS: provided original idea and drafting of the genetic construction, and performed structural analysis experiments by CD spectroscopy. AMMD: provided original ideas and performed the genetic construction and review of the manuscript. and DGR: performed the genetic construction and protein expression experiments. MBR: provided original ideas, study designs, analysis and interpretation of data, drafting and review of the manuscript, graphic and statistical processing and execution of antigenicity, immunogenicity and cellular response studies. IGM and CCHA: performed antigenicity, immunogenicity studies and analysis of cellular response in mouse, rat, and NHP. OCS: study design and supervision of the microneutralization experiments. GLP: contributed to analytical procedures and donor patients selection and evaluation of the immunological and functional response. JVH, EMD, EPV and MAA: study concept and supervision.
Conflict of Interest
MLF, MBR, AMMD, DGR, ACR, GCS, GMP, EPV, MAA and GGN are co-authors of a patent application submitted by the Center for Genetic Engineering and Biotechnology, comprising the C-RBD-H6 PP protein as a vaccine antigen against SASR-CoV-2. All authors approved the final article.
Acknowledgments
The authors would like to acknowledge the assistance provided by Dr. Gabriel Padron and Dr. Diana Garcia del Barco Herrera during the writing of this manuscript. We also thank Dr. Gertrudis Rojas, Dr. Tays Hernández and Dr. Belinda Sanchez from the Center for Molecular Immunology for the recombinant hFc-ACE2, mFc-ACE2 and hFc-RBD chimeric proteins. The authors are especially grateful for technical support to Isela M. Garcia, Yahima Chacón, Ricardo U. Martinez, Dionne Casillas, Ailyn Ramón, Lizet Aldana, Jorge Castro, Juliet M. Enriquez, Mireida Rodríguez, Enrique Noa, Nibaldo L. González and Marta Dubed.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.nbt.2022.08.002.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
References
- 1.Kodati B., Darbha S. Mammalian expression system and improvisation for high production. Int J Sci Res. 2016;5(6):743–749. doi: 10.21275/v5i6.NOV164260. [DOI] [Google Scholar]
- 2.Barone P.W., Wiebe M.E., Leung J.C., Hussein I.T.M., Keumurian F.J., Bouressa J., et al. Viral contamination in biologic manufacture and implications for emerging therapies. Nat Biotechnol. 2020;38(5):563–572. doi: 10.1038/s41587-020-0507-2. [DOI] [PubMed] [Google Scholar]
- 3.Arbeitman C.R., Auge G., Blaustein M., Bredeston L., Corapi E.S., Craig P.O., et al. Structural and functional comparison of SARS-CoV-2-spike receptor binding domain produced in Pichia pastoris and mammalian cells. Sci Rep. 2020;10(1) doi: 10.1038/s41598-020-78711-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karbalaei M., Rezaee S.A., Farsiani H. Pichia pastoris: a highly successful expression system for optimal synthesis of heterologous proteins. J Cell Physiol. 2020;235(9):5867–5881. doi: 10.1002/jcp.29583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Premkumar L., Segovia-Chumbez B., Jadi R., Martinez D.R., Raut R., Markmann A., et al. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci Immunol. 2020;5(48) doi: 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grant O.C., Montgomery D., Ito K., Woods R.J. Analysis of the SARS-CoV-2 spike protein glycan shield reveals implications for immune recognition. Sci Rep. 2020;10(1):14991. doi: 10.1038/s41598-020-71748-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lam J.S., Mansour M.K., Specht C.A., Levitz S.M. A model vaccine exploiting fungal mannosylation to increase antigen immunogenicity. J Immunol. 2005;175(11):7496–7503. doi: 10.4049/jimmunol.175.11.7496. [DOI] [PubMed] [Google Scholar]
- 9.Chen W.H., Pollet J., Strych U., Lee J., Liu Z., Kundu R.T., et al. Yeast-expressed recombinant SARS-CoV-2 receptor binding domain RBD203-N1 as a COVID-19 protein vaccine candidate. Protein Expr Purif. 2022;190 doi: 10.1016/j.pep.2021.106003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen W.H., Tao X., Agrawal A.S., Algaissi A., Peng B.H., Pollet J., et al. Yeast-expressed SARS-CoV recombinant receptor-binding domain (RBD219-N1) formulated with aluminum hydroxide induces protective immunity and reduces immune enhancement. Vaccine. 2020;38(47):7533–7541. doi: 10.1016/j.vaccine.2020.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen W.H., Wei J., Kundu R.T., Adhikari R., Liu Z., Lee J., et al. Genetic modification to design a stable yeast-expressed recombinant SARS-CoV-2 receptor binding domain as a COVID-19 vaccine candidate. Biochim Biophys Acta Gen Subj. 2021;1865(6) doi: 10.1016/j.bbagen.2021.129893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalvie N.C., Biedermann A.M., Rodriguez-Aponte S.A., Naranjo C.A., Rao H.D., Rajurkar M.P., et al. Scalable, methanol-free manufacturing of the SARS-CoV-2 receptor-binding domain in engineered Komagataella phaffii. Biotechnol Bioeng. 2022;119(2):657–662. doi: 10.1002/bit.27979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pollet J., Chen W.H., Versteeg L., Keegan B., Zhan B., Wei J., et al. SARS-CoV-2 RBD219-N1C1: a yeast-expressed SARS-CoV-2 recombinant receptor-binding domain candidate vaccine stimulates virus neutralizing antibodies and T-cell immunity in mice. bioRxiv. 2021 doi: 10.1101/2020.11.04.367359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Espinosa L.A., Ramos Y., Andujar I., Torres E.O., Cabrera G., Martin A., et al. In-solution buffer-free digestion allows full-sequence coverage and complete characterization of post-translational modifications of the receptor-binding domain of SARS-CoV-2 in a single ESI-MS spectrum. Anal Bioanal Chem. 2021;413(30):7559–7585. doi: 10.1007/s00216-021-03721-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med. 2020;383(27):2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Logunov D.Y., Dolzhikova I.V., Shcheblyakov D.V., Tukhvatulin A.I., Zubkova O.V., Dzharullaeva A.S., et al. Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: an interim analysis of a randomised controlled phase 3 trial in Russia. Lancet. 2021;397(10275):671–681. doi: 10.1016/S0140-6736(21)00234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grote A., Hiller K., Scheer M., Munch R., Nortemann B., et al. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005:W526–W531. doi: 10.1093/nar/gki376. 33(Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bai J., Swartz D.J., Protasevich I.I., Brouillette C.G., Harrell P.M., Hildebrandt E., et al. A gene optimization strategy that enhances production of fully functional P-glycoprotein in Pichia pastoris. PLoS One. 2011;6(8) doi: 10.1371/journal.pone.0022577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao X., Huo K.K., Li Y.Y. Synonymous codon usage in Pichia pastoris. Sheng Wu Gong Cheng Xue Bao. 2000;16(3):308–311. [PubMed] [Google Scholar]
- 20.Higgins D.R., Busser K., Comiskey J., Whittier P.S., Purcell T.J., Hoeffler J.P. Small vectors for expression based on dominant drug resistance with direct multicopy selection. Methods Mol Biol. 1998;103:41–53. doi: 10.1385/0-89603-421-6:41. [DOI] [PubMed] [Google Scholar]
- 21.Gurramkonda C., Polez S., Skoko N., Adnan A., Gabel T., Chugh D., et al. Application of simple fed-batch technique to high-level secretory production of insulin precursor using Pichia pastoris with subsequent purification and conversion to human insulin. Micro Cell Fact. 2010;9:31. doi: 10.1186/1475-2859-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Betancourt L.H., Espinosa L.A., Ramos Y., Bequet-Romero M., Rodriguez E.N., Sanchez A., et al. Targeting the hydrophilic regions of recombinant proteins by MS via in-solution buffer-free trypsin digestion. Eur J Mass Spectrom. 2020;26(3):230–237. doi: 10.1177/1469066719893492. [DOI] [PubMed] [Google Scholar]
- 23.Blanco O.R., Dorta D., Hernandez C.A., Abreu D., Dominguez A.G., Luna Y., et al. Murine monoclonal antibodies against RBD of the SARS-CoV-2 spike protein as useful analytical tools for subunit vaccine development and clinical trials. J Immunol Methods. 2022;500 doi: 10.1016/j.jim.2021.113195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181(4) doi: 10.1016/j.cell.2020.03.045. 894-904 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183(4) doi: 10.1016/j.cell.2020.09.037. 1024-1042 e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584(7821):437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suthar M.S., Zimmerman M.G., Kauffman R.C., Mantus G., Linderman S.L., Hudson W.H., et al. Rapid generation of neutralizing antibody responses in COVID-19 patients. Cell Rep Med. 2020;1(3) doi: 10.1016/j.xcrm.2020.100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Rosmalen M., Krom M., Merkx M. Tuning the flexibility of glycine-serine linkers to allow rational design of multidomain proteins. Biochemistry. 2017;56(50):6565–6574. doi: 10.1021/acs.biochem.7b00902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dalvie N.C., Rodriguez-Aponte S.A., Hartwell B.L., Tostanoski L.H., Biedermann A.M., Crowell L.E., et al. Engineered SARS-CoV-2 receptor binding domain improves manufacturability in yeast and immunogenicity in mice. Proc Natl Acad Sci USA. 2021;118(38) doi: 10.1073/pnas.2106845118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malladi S.K., Singh R., Pandey S., Gayathri S., Kanjo K., Ahmed S., et al. Design of a highly thermotolerant, immunogenic SARS-CoV-2 spike fragment. J Biol Chem. 2021;296 doi: 10.1074/jbc.RA120.016284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pino M., Abid T., Pereira Ribeiro S., Edara V.V., Floyd K., Smith J.C., et al. A yeast expressed RBD-based SARS-CoV-2 vaccine formulated with 3M-052-alum adjuvant promotes protective efficacy in non-human primates. Sci Immunol. 2021;6(61) doi: 10.1126/sciimmunol.abh3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grabovenko F., Nikiforova L., Yanenko B., Ulitin A., Loktyushov E., Zatsepin T., et al. Glycosylation of receptor binding domain of SARS-CoV-2 S-protein influences on binding to immobilized DNA aptamers. Int J Mol Sci. 2022;23(1) doi: 10.3390/ijms23010557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zang J., Zhu Y., Zhou Y., Gu C., Yi Y., Wang S., et al. Yeast-produced RBD-based recombinant protein vaccines elicit broadly neutralizing antibodies and durable protective immunity against SARS-CoV-2 infection. Cell Disco. 2021;7(1):71. doi: 10.1038/s41421-021-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duman J.G., Miele R.G., Liang H., Grella D.K., Sim K.L., et al. O‐Mannosylation of Pichia pastoris cellular and recombinant proteins. Biotechnol Appl Biochem. 1998;28(1):39–45. doi: 10.1111/j.1470-8744.1998.tb00510.x. [DOI] [PubMed] [Google Scholar]
- 35.Trimble R.B., Lubowski C., Hauer C.R., 3rd, Stack R., McNaughton L., et al. Characterization of N- and O-linked glycosylation of recombinant human bile salt-stimulated lipase secreted by Pichia pastoris. Glycobiology. 2004;14(3):265–274. doi: 10.1093/glycob/cwh036. [DOI] [PubMed] [Google Scholar]
- 36.Autenrieth S.E., Autenrieth I.B. Variable antigen uptake due to different expression of the macrophage mannose receptor by dendritic cells in various inbred mouse strains. Immunology. 2009;127(4):523–529. doi: 10.1111/j.1365-2567.2008.02960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luong M., Lam J.S., Chen J., Levitz S.M. Effects of fungal N- and O-linked mannosylation on the immunogenicity of model vaccines. Vaccine. 2007;25(22):4340–4344. doi: 10.1016/j.vaccine.2007.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maciola A.K., Pietrzak M.A., Kosson P., Czarnocki-Cieciura M., Smietanka K., et al. The length of N-glycans of recombinant H5N1 hemagglutinin influences the oligomerization and immunogenicity of vaccine antigen. Front Immunol. 2017;8:444. doi: 10.3389/fimmu.2017.00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 40.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581(7807):221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sui J., Deming M., Rockx B., Liddington R.C., Zhu Q.K., et al. Effects of human anti-spike protein receptor binding domain antibodies on severe acute respiratory syndrome coronavirus neutralization escape and fitness. J Virol. 2014;88(23):13769–13780. doi: 10.1128/JVI.02232-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D’Alessio C., Caramelo J.J., Parodi A.J. UDP-GlC: glycoprotein glucosyltransferase-glucosidase II, the ying-yang of the ER quality control. Semin Cell Dev Biol. 2010;21(5):491–499. doi: 10.1016/j.semcdb.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material