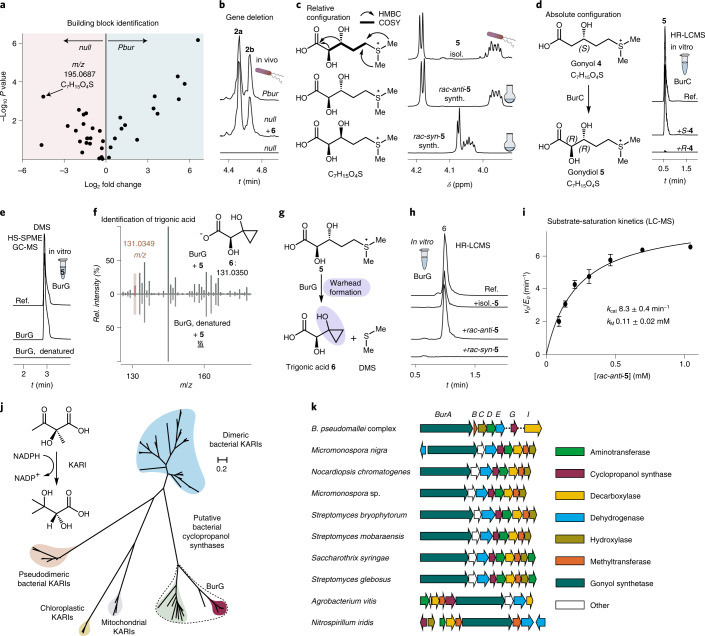
Fig. 2. Substrate identification and reconstitution of enzymatic cyclopropanol formation.
a, Metabolomics analysis of B. thailandensis Pbur versus B. thailandensis Pbur BurG::Kan (malleicyprol null mutant) cell extracts (biological triplicates, n = 3). Data filtered for sulfur-containing compounds; see Supplementary Fig. 1 for non-filtered analysis. b, Chemical complementation of the null mutant with 6 restores malleicyprol production (isoforms 2a and 2b) to wild-type level. c, Structure elucidation of isolated (isol.) gonydiol 5, and comparison of key 1H NMR shifts with synthetic (synth.) racemic syn- and anti-isoforms. COSY, correlated spectroscopy; HMBC, heteronuclear multiple bond correlation. d, Formation of 5 catalysed by hydroxylase BurC in assays with addition of R-4 or S-4; top trace: reference (5). e, Detection of DMS as product of BurG reactions by GC-MS. f, Mirror plot of enzymatic assay (top) of BurG incubated with 5 versus denatured BurG (bottom) indicates a compound with the m/z value of 6 as enzymatic product; see Extended Data Fig. 3 for chromatogram. g, BurG-catalysed transformation of 5 to 6. h, Monitoring of enzymatic turnover of isolated 5 and synthetic rac-syn-5 or rac-anti-5 into 6 via HR-LCMS. i, Steady-state kinetics of formation of 6 from rac-anti-5; mean and standard deviation obtained from two independent preparations (n = 2). j, Phylogenetic analysis of BurG, orthologues and canonical KARIs. The scale bar indicates amino acid substitutions per site. k, Alignment of biosynthetic gene cluster encoding for BurG orthologues clustered with the genes associated with biosynthesis of 5.