Fig. 2. Assembly and disassembly of the DNA-based filaments in cell-sized confinement.
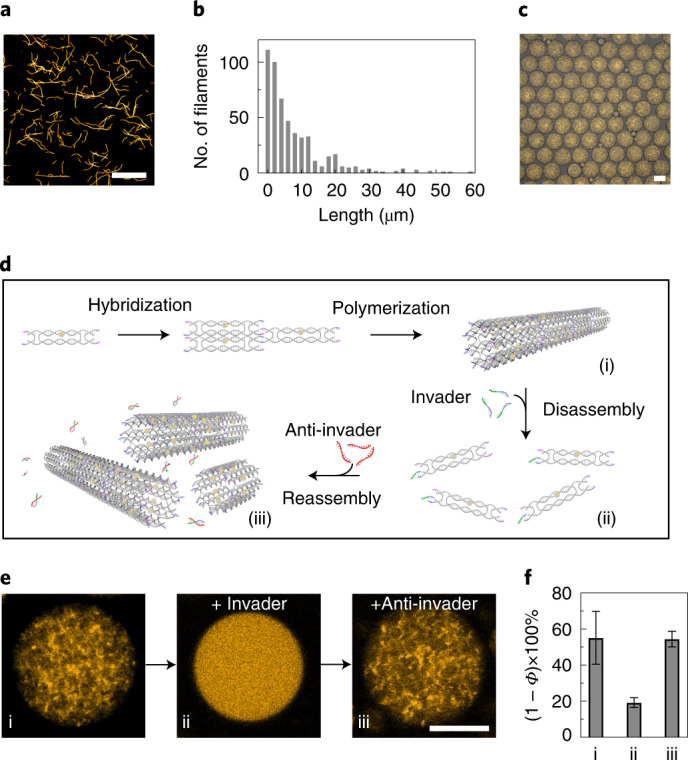
a, Confocal microscopy image of the Cy3-labelled DNA-based filaments (excitation wavelength, λex = 561 nm). Scale bar, 20 μm. b, Histogram of the filament lengths determined by confocal microscopy, showing a mean length of 7.74 μm (n = 516). c, Overlay of the confocal and bright-field overview images of the monodisperse microfluidic water-in-oil droplets containing Cy3-labelled DNA-based filaments (excitation wavelength, λex = 561 nm). Scale bar, 50 μm. d, Schematic of the DNA tile design with toeholds13. Addition of the invader strands leads to the disassembly of the filaments, whereas addition of the anti-invader strands leads to reassembly. e, Representative confocal images of the DNA-based filaments encapsulated into droplets (i) before and (ii) after the addition of the invader strands and (iii) after the addition of the anti-invader strands. Upon addition of the invader strands the filaments are disassembled, leading to a homogeneous distribution of the fluorescence signals inside the droplet. Scale bar, 20 μm. f, Histogram of the porosity ((1 − Φ) × 100%, reflecting the degree of polymerization) of the DNA-based filaments encapsulated into water-in-oil droplets (i) in the absence, (ii) in the presence of the invader strands (10 μM) and (iii) after addition of the anti-invader strands (37.5 μM). Error bars correspond to the standard deviation of n ≥ 5 droplets.
