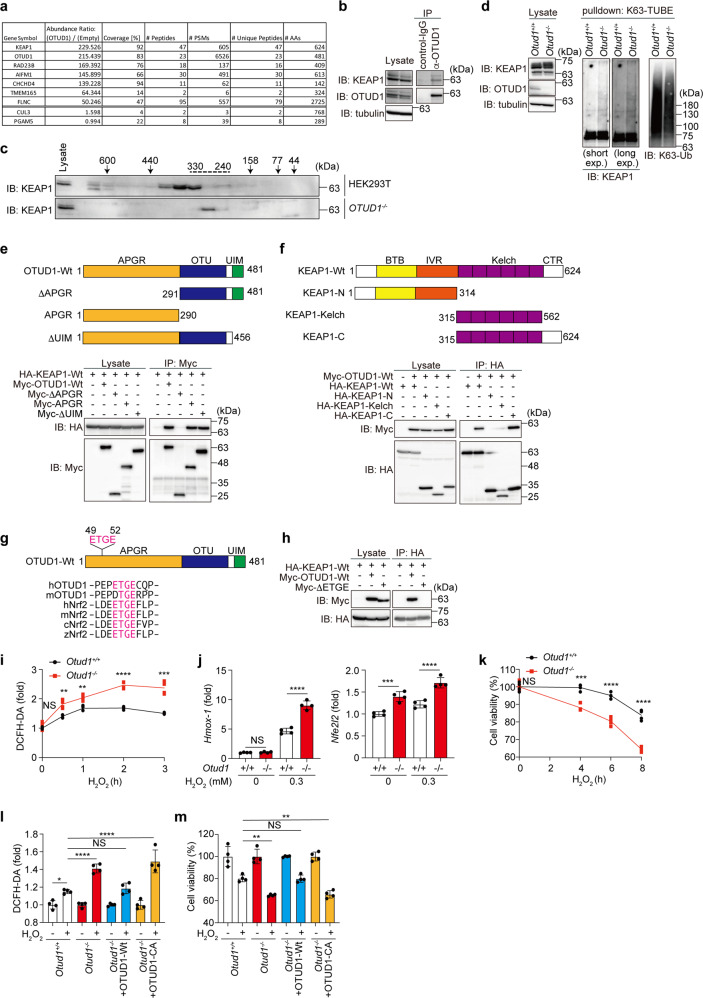
Fig. 5. OTUD1 regulates KEAP1-mediated oxidative stress response and ROS-associated cell death.
a Identification of OTUD1-interacting proteins. FLAG-tagged OTUD1 was expressed in HEK293T cells and immunoprecipitated with an anti-FLAG antibody, and the co-immunoprecipitants were then analyzed by MS. Proteins with an abundance ratio >50, and values for CUL3 and PGAM5 are shown. b OTUD1 physiologically interacts with KEAP1. HEK293T cell lysates and anti-OTUD1 immunoprecipitates were subjected to immunoblotting with the indicated antibodies. c KEAP1 in OTUD1−/− cells eluted at a lower molecular weight as compared to that in parental cells. Gel filtration fractions from parental and OTUD1−/− cells, as shown in Fig. 1f, were immunoblotted with an anti-KEAP1 antibody. d OTUD1 regulates K63- ubiquitination of KEAP1. Cell lysates and K63-TUBE precipitates from Otud1+/+- and Otud1−/−-MEFs were immunoblotted with the indicated antibodies. e APGR of OTUD1 is required for KEAP1-binding. Wt- and mutants of Myc-OTUD1 were expressed with HA-KEAP1 in HEK293T cells. Cell lysates and anti-Myc-immunoprecipitates were immunoblotted with the indicated antibodies. f The Kelch domain and the C-terminal region (CTR) of KEAP1 is responsible for OTUD1-binding. A similar analysis as in e was performed using Wt- and mutants of HA-KEAP1 and Myc-OTUD1. g OTUD1 contains an ETGE motif in APGR. Localization of the ETGE motif in OTUD1, and amino acid sequence alignment with NRF2 are shown [26]. h: human, m: mouse, c: chicken, z: zebrafish. h The ETGE motif in OTUD1 is indispensable for KEAP1-binding. Myc-tagged Wt- or ETGE motif-deleted mutant of OTUD1 was expressed with HA-KEAP1-Wt as indicated, and cell lysates and anti-HA immunoprecipitates were immunoblotted with the depicted antibodies. i Enhanced hydrogen peroxide-induced ROS generation in Otud1-deficient cells. Otud1+/+- and Otud1−/−-MEFs were treated with 0.3 mM H2O2 for the indicated periods, and the intracellular ROS levels were analyzed by a DCFH-DA assay. j Enhanced expression of NRF2 target genes in Otud1−/−-MEFs. MEFs were treated with or without 0.3 mM H2O2 for 3 h, and qPCR analyses were performed. k Reduced cell viability in Otud1−/−-MEFs under oxidative stress. MEFs were treated with 0.3 mM H2O2 for the indicated periods, and cell viability was analyzed by Celltiter Glo assay. l DUB activity of OTUD1 is necessary to suppress ROS production. OTUD1-Wt or active site mutant (CA) was restored into Otud1−/−-MEFs, and ROS levels were analyzed by a DCFH-DA assay after treatment with or without 0.3 mM H2O2 for 2 h. m DUB activity of OTUD1 protects cells from H2O2-induced death. A similar treatment as in l was performed with or without 0.1 mM H2O2 for 8 h, and cell viability was analyzed by a Celltiter Glo assay. Data are shown as mean ± SD by t-test of each time point (n = 4, i, k) or ANOVA post-hoc Tukey test (n = 4, j, l, m). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, NS not significant.