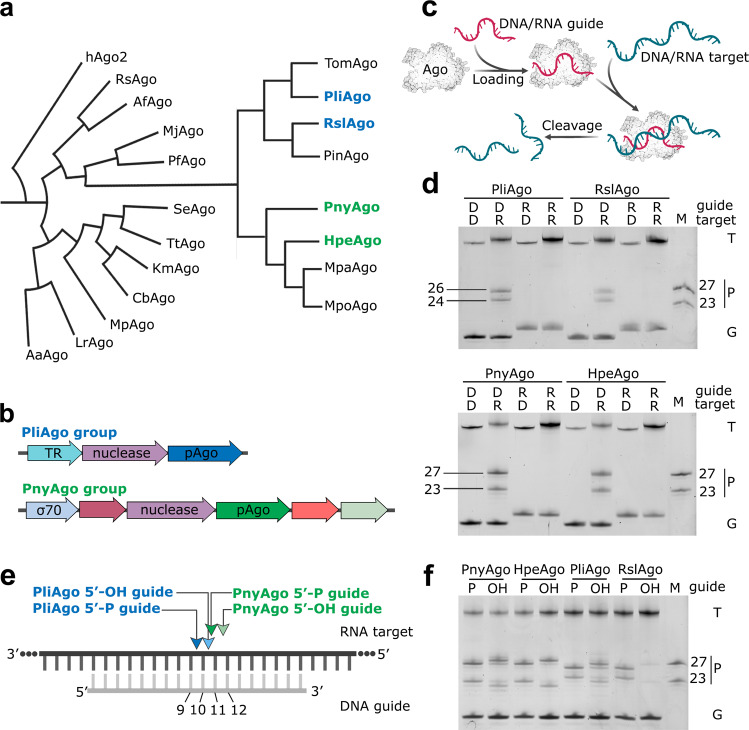
Fig. 1. Two groups of pAgos prefer DNA guides and RNA targets.
a Phylogenetic tree of previously studied pAgos and of predicted D-R pAgos; pAgos studied in this work are shown in color (see Supplementary Fig. 1a for the full list of D-R pAgos and abbreviations). b Operon structure of D-R pAgos from the PliAgo (top) and PnyAgo (bottom) groups. pAgo-associated nuclease from the PD-(D/E)XK superfamily, putative transcription regulator (TR) and σ70 family factor are indicated. c Scheme of the cleavage assay. d Nucleic acid specificity of the two groups of pAgos. The reactions were performed with DNA (‘D’) or RNA (R’) guides and targets; positions of targets (‘T’), guides (‘G’) and reaction products (‘P’) are indicated (SYBR Gold staining). e Schematics of target RNA cleavage by the two groups of D-R pAgos in the presence (dark arrowheads) and in the absence (light arrowheads) of the 5’-phosphate in guide DNA. Nucleotide positions in guide DNA are indicated. f Analysis of RNA cleavage by D-R pAgos with 5′-P and 5′-OH guide DNAs. In panels d and f representative gels from 3 independent experiments with similar results are shown.