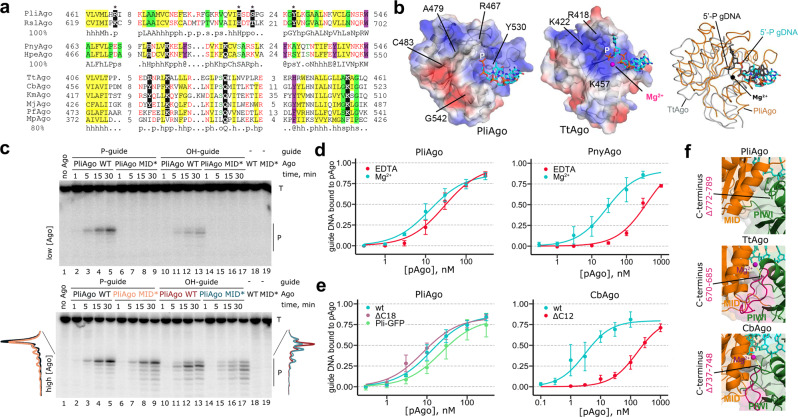
Fig. 3. Role of the MID pocket in guide DNA binding and target RNA cleavage.
a Alignment of the MID pocket in various groups of pAgos. Conserved residues of the MID motif involved in 5’-nucleotide interactions are indicated in black. MpAgo has a hydrophobic MID pocket and binds 5′-hydroxylated guide RNAs but is more closely related to canonical pAgos than to PliAgo. The consensus for each group is shown underneath the alignment: ‘h’, hydrophobic residues (WFYMLIVACTH); ‘p’, polar (EDKRNQHTS); ‘s’, small (ACDGNPSTV); ‘o’, OH-containing (ST); ‘@‘, aromatic (YWFH). b Interactions of the MID domain with guide 5’-end (stick) in PliAgo (left) and TtAgo (PDB: 3DLH32) (middle). Electrostatic potentials are shown as surfaces (blue, basic; red, acidic; white, neutral). (Right) Overlay of the MID pockets of PliAgo/P-gDNA (MID, orange; DNA, cyan) and TtAgo/gDNA complexes (MID, gray; DNA and Mg2+, black). c Kinetics of RNA cleavage by wild-type and MID* PliAgo (R467A/Y530A) loaded with 5’-P or 5’-OH gDNA, at low (50 nM) or high (500 nM) PliAgo concentrations (representative gel from two independent experiments). The reactions were performed with 5’-P32-labeled target RNA; positions of the target (‘T’) and the reactions products (‘P’) are indicated. The profiles of the cleavage products observed after 30 min with 500 nM PliAgo are shown on the left for P-gDNA (WT PliAgo, black; MID* PliAgo, orange) and on the right for OH-gDNA (WT, red; MID*, cyan). d Guide DNA binding by PliAgo and PnyAgo in the presence and in the absence of Mg2+. e Guide DNA binding by PliAgo, CbAgo and their variants PliAgo ΔC18 (deletion of the C-terminal 18 residues), PliAgo-GFP (C-teminal fusion with GFP), CbAgo ΔC12 (deletion of the C-terminal 12 residues). In d and e, means from 3 or 4 independent experiments are shown; error bars correspond to standard deviations. The amount of bound DNA is shown as a fraction of total 5’-P32-labeled DNA in the sample. f Comparison of the C-terminus positions in the structures of PliAgo (top), TtAgo (3DLH, middle) and CbAgo (6QZK, bottom). The C-terminal segment deleted in PliAgo (Δ772-789) is not solved on the structure. The corresponding segments in TtAgo (670-685) and CbAgo (737–748) are shown in magenta based on structural alignment.